This part of the Forsburg Lab website contains technical information of use to people who study S. pombe. Visit our home page for a directory to this pombe site, or the list of frequently asked questions for fast answers to common questions. If you want to browse practical information about working with fission yeast, you’re in the right place.
On this page
- Commonly used selectable markers.
- A note about nomenclature.
- A summary of promoter activity and expression systems
- Info on primer design for amplification of open reading frames.
- How to construct plasmids for cross-complementation experiments
Other pages on our site: follow these links to find
- Frequently asked questions about working with pombe
- Almanac of useful facts, ade6 alleles, restriction sites, codon usage
- PombeWeb, including fission yeast lab home pages, faculty listings, meetings, genomics, and all the pombe-related links we can find.
- An index and table of fission yeast plasmids, including general plasmid information, sequences, and maps (the vector database).
- A list of useful older technical references for fission yeast molecular genetics.
- Sequence analysis sites including Sanger, the Blast servers and analysis tools
- Recipes for fission yeast media
- Drugs for fission yeast, including selection/counterselection and DNA damaging agents
- Want to know the equivalent S. cerevisiae genes for S. pombe cell cycle genes? Visit our gene conversion table.
- Where to get strains and plasmids.
- G1 phase? check out the cell cycle page to figure out where G1 went
Common S. Pombe Markers
- ade1+ complements ade1
- ade6+ complements ade6
- arg3+ complements arg3
- CAN1 from S. cerevisiae complements can1 , which encodes an arginine permease. can1+ cells are sensitive to canavanine, a toxic arginine analogue; this is a counterselectable marker. Note that can1 his3 and can1 his7 double mutants have a slow growth phenotype, presumably due to the overlapping histidine permease activity of the can1+ gene.
- his3+ complements his3
- his7+ complements his7
- leu1+ complements leu1
- LEU2 from S. cerevisiae complements leu1; growth is slow in single copy
- sup3-5, a nonsense suppressor, rescues ade6-704. Useful to select for single copy integrants. ade6-704 colonies are dark red in limiting adenine. In the presence of the sup3-5 marker on a plasmid, which is slightly toxic in high copy, they are pink due to plasmid loss. If the sup3-5 marker integrates, it becomes stable and the colonies are white.
- ura4+ complements ura4
- URA3 from S. cerevisiae complements ura4 but very weakly
- Drug markers: G418 (kanMX), hygromycin (hyp), Clonnat/noursethecin (nat). These can be used in YE/YES media or PMG, but not EMM.
What’s in a name?
Naming a new gene? It’s a good idea to check at the Sanger Centre pombe genome project to see whether your gene designation is already taken. You can also reserve a gene name for a particular locus prior to publication. Investigators are urged to use the gene registry to reserve names, to propose name changes, or to contact the GNC (gene nomenclature committee). The GNC will address duplicate names, non-standard designations, and other such issues. It will also revise the nomenclature rules as needed to ensure that they are up to date. Access the committee through the site above.
General nomenclature rules: Fission yeast gene names are generally expressed as a three letter, italic name followed by a number, and a plus for wild type (cdc19+; the plus should be superscripted in normal text). In mutants, the plus is replaced by an allele designation (cdc19-P1). Allele designations vary widely in format; some are letters, some numbers, some a mix.
Most investigators indicate deletions with a Δ(delta). For example, a disruption of your favorite gene yfg1+ with ura4+ is written Δyfg1::ura4+ in a proper strain table, although people often use Δ yfg1 or yfg1Δ as short hand in text. Recently, yfg1Δ has become the preferred designation. For deletion alleles constructed without an insertion, some use an allele designation beginning with D; e.g, ura4-D18 is a deletion of ura4+. However, you cannot be sure that a “D” in an allele designation is always a deletion, so use caution and read the literature. It is strongly encouraged by some that deletion alleles be given a unique allele designation, e.g., yfg1-D1::ura4+ but this isn’t typical. However, in several cases, the same gene has been disrupted in different laboratories, and the phenotypes vary according to the precise construct. Without a unique identifier for a given lab’s allele, this has led to considerable confusion in the literature.
The :: (double colon) is used to indicate an insertion into the genome. It need not correspond to a deletion/disruption, for example, marking a locus by integrating a marker downstream would be indicated yfg1+::leu1+ or yfg1+::pAB123. Generally, what comes before the :: is the locus, and you need to indicate whether it is wild type or mutant; what comes after the :: is what was integrated, and you need to identify it as well. In some cases, people use a single colon : to indicate an insertion, and a double colon :: to indicate a replacement/disruption.
Proteins are indicated by roman text. In older literature, this was followed by an appended p (eg, Cdc19p). The p is particularly useful for non-yeast people who might be unfamiliar with the conventions of roman versus italic type, but it has fallen out of favor. Just to complicate life further, budding yeast nomenclature is different: the wildtype gene names are in italic capitals (LEU2), mutants (unless dominant) are in lower case (leu2-3) and protein names are in roman text (LEU2 or Leu2).
Expression Vectors
A number of cloned promoters in fission yeast for expression of heterologous genes have been characterized. They are adh1+ (constitutive high expression), fbp1+ (carbon source responsive), a tetracycline-repressible system based on the CaMV promoter, and the nmt1+ (no message in thiamine) promoter, which is the most frequently used. A newer promoter with a quick induction is the urg1+ promoter, which is induced by the addition of uracil to the media, although it doesn’t work well outside of its normal locus. There is a copper-based promoter ctr4+ which is induced by removing copper from the media, and the inv1+ invertase promoter, which undergoes transient induction by sucrose. References for these can be found on our technical references page.
By far the most common expression system is nmt1+. There are three versions of nmt1+ promoter: the full strength promoter, and two attenuated versions that have reduced activity both in repressed and induced conditions (indicated below as nmt* /nmt41, and nmt**/nmt81; see references). Several different polylinkers are available in the REP/RIP series of nmt vectors (see vector database). The concentration of thiamine can be adjusted for partial activation. Full induction: no thiamine. Full repression: 15uM thiamine (5ug/ml). Partial induction (described in this Javerzat et al, Nucl. Acids Res. 24: 4676-4683 ): 0.05uM thiamine (0.016ug/ml).
The nmt1 promoter does not switch off completely, and the ability to construct a “shut-off” plasmid depends very much on the protein being expressed and the sensitivity of the cell to dosage of that particular protein. Many genes expressed under nmt1 control are able to complement even in the presence of thiamine in the weakest promoter, but there are also numerous examples of genes that can be successfully shut off to generate a null phenotype. Thus, the utility of this promoter for plasmid shut-off experiments must be determined empirically for each gene.
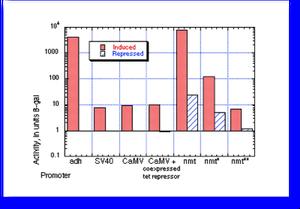
A comparison of promoter activity for the older promoters was published in Forsburg, (1993). Nucl. Acids Res. 21, 2955-2956. The data are summarized as in the figure (measuring units of beta galactosidase activity produced by lacZ fusions). The first three are constitutive, although the CaMV promoter can be rendered inducible by coexpression of a tet repressor protein and we have a strain that has this integrated (see vectors page).
Primer Design
From Juerg Bahler: Web-interface scripts that automatically design primers for PCR-based gene targeting. This will automatically suggest primer sequences for gene deletion, tagging, and/or regulatable expression based on gene name and plasmid information that you specify.
Plasmid Shuffle
There are at least four systems that have been used. The classic 5-FOA counterselection against ura4+ works well in our hands (e.g., Liang 2001), although there can be background problems in some strains. A recipe for FOA media is on our drugs page. Another method, developed by Moser et al., uses a color based system and the ade1+ gene. A third uses the CAN1 plasmids developed for this purpose by Ekwall and Ruusala and was reported by Paluh and Clayton. Finally, expressing a viral thymidine kinase gene as the counterselectable marker allows you to use FUDR (a toxic thymidine analogue) instead of FOA. This isn’t quite as fussy as FOA–see the method in Kiely et al–and has the additional advantage of not requiring any particular markers in the chromosome.
We have had success generating ts alleles using plasmid shuffle strategies in pombe. Although written for cerevisiae, this protocol for isolation of ts mutants more or less explains how it’s done. Our method: we isolate a haploid containing a disruption of our favorite gene yfg1+, covered by a ura4+-marked (or tk-marked) plasmid containing the wild type gene (call it plasmid 1). We use hydroxylamine for in vitro mutagenesis of another yfg1+ plasmid, marked with something else (plasmid 2). We transform plasmid 2 into the starting haploid, use plasmid shuffle with FOA or FUDR selection against plasmid 1, and screen for ts mutants that contain plasmid 2 and lack plasmid 1. Upon isolation and retesting of the plasmid to confirm the temperature-sensitive phenotype, we integrate back into the chromosomal locus.
Charlie Hoffman (Pombe2011 meeting) notes that FOA in fission yeast induces mutation in the ura5+ gene as well; this isn’t a problem in budding yeast where there are two URA5 homologues. However, it does mean that in pombe, there will be a background of FOA-resistant cells that maintain the original ura4+.