This procedure is typically used for the isolation and preparation of spores from a diploid strain heterozygous for a marked disruption (e.g., yfg1::his3+) Inoculation of the spore population into minimal medium lacking the nutritional supplement corresponding to the disruption marker (e.g., minimal medium lacking histidine) allows only the disruption spores to germinate. In this example, the wild type spores are histidine auxotrophs and are unable to germinate or enter the cell cycle. (We have also successfully used ura4+ and LEU2 markers in these assays.) By following the population using FACS and microscopy, one can determine whether the disrupted cells succeed in synthesizing DNA and arrest in the first cell cycle. Versions of this method have been published in several papers; this is the protocol in use in the Forsburg lab and was derived from that described by Kelly et al (1993).
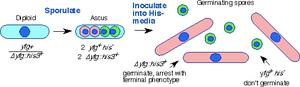
Diagram of spore germination procedure. In this case, an essential gene yfg1+ has been disrupted with his3+ to generate a heterozygous diploid. The diploid is induced to sporulate, the spores are isolated in a large scale random spore analysis, and then inoculated in bulk into media that selects for histidine prototrophy. Only the yfg1::his3 disruptants will germinate, and they can be distinguished from the wild type on the basis of morphology (the wild type spores will remain inert) and by FACS (the wild type spores will remain at 1C).
Method
Before beginning: verify that your diploids are spo+! Streak onto YE + phloxin B, and then replica onto YES and ME. You can check for spo+ diploids after two days, and then pick a colony from the YES plate. Also, this protocol can use a lot of glusalase. Check now to make sure there will be enough!
- Inoculate a spo+ diploid colony and grow to OD(595) 0.8 – 0.9 in YES at 32*C. (25*C if ts diploid)
- Inoculate 10 – 15 mls of YES culture into 200 mls filtered ME broth. Grow at 25*C for three or four days.
- Check microscopically to make sure that the cells have sporulated nicely.
- Harvest spores/asci and resuspend in 20 mls of 2% glusalase overnight at 25*C (150 rpm).
- Check microscopically that asci and vegetative cells have been digested.
- In 50 ml Falcons, wash five times with 15 mls of rinsing media. Vortexing is okay.
- In a fresh 50 ml Falcon prepare 40 mls of 25% glycerol (made by mixing sterile 100% glycerol and rinsing media) for each strain.
- Add 5 mls of rinsing media to harvested pellets. Vortex into suspension.
- Layer spores on top of glycerol.
- Centrifuge 10 – 15 mintues at 2000-3000 rpm.
- Remove sup and save for now in case you lose your spores (rare).
- Wash pellet 3 times with 15 mls of rinsing media.
- Resuspend pellet in 10 ml of media.
- Examine microscopically to make sure prep is clean.
- Plate small amount on YES to make sure phenotypes are correct .
- Store spores at 4*C.
- Inoculate spores into EMM-selective medium at 107 to 108 spores per mL. To carry out a long timecourse, you can set up staggered cultures–one in the morning and one in the evening. Make sure you treat them exactly the same and have several timepoints overlapping (eg, 9 and 10 hours), so that you can be sure they are behaving identically.
Notes
“clean” is a relative term–you can never get rid of all of the dead vegetative cells. If there is a lot of junk remaining in your prep you can try to clean it up by spinning through another glycerol cushion. Otherwise you can judiciously gate out the small stuff when you plot your FACS. Note that checkpoint defective strains generate small aploid cells, so be careful when gating that you don’t lose real data!
Rinsing media = Yeast Nitrogen Base – Ammonium Sulfate = 1.7 g powder/L MQ water; filter sterilize or autoclave.
Important: keep spores free of glucose to keep them from germinating prematurely!