Activation of the MCM complex depends on the DDK kinase, which comprises the kinase subunit Hsk1/Cdc7, and a regulatory subunit Dfp1/Dbf4. Like a cyclin, Dfp1 targets the kinase to its substrates, and without it, Hsk1 appears to be ineffecdtive. Dfp1 is regulated both transcirptionally and post-translationally to restrict kinase activity to the G1/S phase of the cell cycle.
Work largely performed in budding yeast (but also by some fission yeast groups) suggests that DDK has an essential function in replication by phosphorylating the MCM proteins. Intriguingly, however, this effect can be bypassed by certain mutations in Mcm4 and Mcm5. This suggests that the base state of the MCMs is inactive, and DDK relieves this inhibition allowing the helicase to function. We showed that DDK activity is required to load the replication helicase activator, Cdc45, on the chromatin.
DDK also is regulated by the Cds1 checkpoint . In fact, Hilary Snaith showed loss of cds1 functions as a genetic suppressor of hsk1-1312, a temperature sensitive allele of hsk1+, which is consistent with Cds1 inhibiting Hsk1 activity. We also observe that Hsk1 undergoes Cds1-dependent phosphroylation.
However, DDK has a number of additional functions. These can in many cases be mapped to specific domains of the Dfp1 subunit, which allows selective inactivation of certain pathways (genetic separation of function mutations). Although originally, people thought DDK was only required for initiation of DNA replication, its functions now appear quite diverse, and are linked primarily by the necessity to carry them out in S phase when the kinase is active. There is evidence from us, as well as the labs of Jacob Dalgaard and Hisao Masai, that DDK functions at the replication fork, with the Fork Protection Complex proteins Swi1 and Swi3.
Finally, we isolated a high copy suppressor of Hsk1, the phosphatase Ibp1. This functions genetically in a pathways separate from Cds1. We are interested in determining how Ibp1 contributes to suppress the effect of hsk1-1312.
DDK and the Centromere: Chromosome Segregation and Replication Timing

Hilary Snaith identified our ts allele of hsk1 and noticed a phenotype she called “fragmented nuclei”. When Julie Bailis joined the lab, she tracked this down to a chromosome segregation defect. Working with Robin Allshire, we identified a role for DDK at the centromere. hsk1 mutants have a phenotype called lagging chromosomes, in which the cells fail to segregate their chromosomes properly. Instead, some chromosomes lag behind on the metaphase, plate, because they have inappropriately attached their chromosomes to both sides of the spindle. Robin’s group identified Dfp1 as a Swi6-associated protein. Dfp1 contains a short sequence called a “MIR” motif that is often found in Swi6-interacting proteins. Interestingly, this motif is not present in the budding yeast ortholog DBF4–which may make sense because budding yeast lacks an orthologue of Swi6!
We showed that loss of DDK activity leads to disruption of centromere cohesion. Thus, DDK activity in S phase influences chromosome segregation in M phase.
Hisao Masukata’s lab showed that DDK also is required for timely centromere replication. In contrast to other heterochromatin domains, the fission yeast centromere replicates early, and Masukata’s group showed that this depends on the recruitment of DDK by Swi6. (You can read more about Swi6 on the chromosome page). We found that Swi6 also binds to the replication protein Cdc18, which is required to load the MCM complex on the chromatin. Cdc18 also contributes to centromere replication timing.
DDK and Alkylating DNA Damage
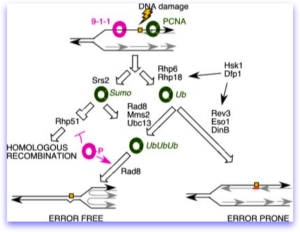 A number of years ago, Grant Brown’s lab made a series of mutations truncating the C-terminus of Dfp1. These all caused the cells to be sensitive to alkylating DNA damage (e.g., MMS). Henning Schmidt performed a screen for MMS-sensitive mutations, some with meiotic phenotypes and isolated an allele of dfp1 that also is a C-terminal truncation. Additional mutations from Henning’s screen include alleles of swi9, the MRN complex, and several uncharacterized genes we haven’t been able to clone.
A number of years ago, Grant Brown’s lab made a series of mutations truncating the C-terminus of Dfp1. These all caused the cells to be sensitive to alkylating DNA damage (e.g., MMS). Henning Schmidt performed a screen for MMS-sensitive mutations, some with meiotic phenotypes and isolated an allele of dfp1 that also is a C-terminal truncation. Additional mutations from Henning’s screen include alleles of swi9, the MRN complex, and several uncharacterized genes we haven’t been able to clone.
Will Dolan observed that when cells are treated with alkylating damage, DDK remains on the chromatin even after S phase. Interestingly, DDK mutations disrupt induced mutagenesis. That is, when treated with a DNA damaging agents, one of the normal cellular responses is to induce activity of error prone DNA polymerases (so-called “sloppier copiers”) in the translesion synthesis, or TLS pathway. These “error prone” and “error free” pathways which depend on highly conserved regulators, are well characterized. Anthony Le found that in fission yeast (as was observed by others in budding yeast), DDK mutant cells have a much lower rate of this induced mutation than wild type. Epistasis analysis places the DDK making a unique input into the TLS pathway, although the mechanism is not clear. However, data in other systems also suggest that it phosphorylates the Rhp18/Rad18 protein, which is part of the master pathway regulator of this DNA damage response.
The dfp1 truncations that cause disruptions in MMS response also disrupt meiosis which we discuss elsewhere.
The diagram is derived from Dolan et al (2010).
Reviews on DDK
- Vaziri C, Masai H. Integrating DNA replication with trans-lesion synthesis via Cdc7. Cell Cycle. 2010 Dec 15;9(24):4818-23.
- Tanaka S, Araki H. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma. 2010 Dec;119(6):565-74.
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010 Jun 15;24(12):1208-19.
Our primary research papers on DDK
- Le, AH, Mastro, T.L, Forsburg SL. (2013) The C-terminus of S. pombe DDK subunit Dfp1 is required for meiosis-specific transcription and cohesin cleavage. Biol Open. 2013 Jun 11;2(7):728-38.
- Dolan WP, Le AH, Schmidt H, Yuan JP, Green M, Forsburg SL. (2010) Fission yeast Hsk1 (Cdc7) kinase is required after replication initiation for induced mutagenesis and proper response to DNA alkylation damage. Genetics. 2010 May;185(1):39-53.
- Dolan WP, Sherman DA, Forsburg SL.Schizosaccharomyces pombe replication protein Cdc45/Sna41 requires Hsk1/Cdc7 and Rad4/Cut5 for chromatin binding.Chromosoma. 2004 Sep;113(3):145-56.
- Bailis JM, Bernard P, Antonelli R, Allshire RC, Forsburg SL.Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat Cell Biol. 2003 Dec;5(12):1111-6.
- Snaith HA, Marlett J, Forsburg SL.Ibp1p, a novel Cdc25-related phosphatase, suppresses Schizosaccharomyces pombe hsk1 ( cdc7). Curr Genet. 2003 Oct;44(1):38-48.
- Snaith, H.A., Brown, G., and Forsburg, S.L. (2000) S. pombe Hsk1p is a potential Cds1p target required for genome integrity. Mol. Cell. Biol. 20:7922-7932. PMC86403