Several features of fission yeast make it a particularly good model for the study of chromosome biology. Fission yeast has a genome of about 12 Mb divided between just three chromosomes. The relatively large fission yeast chromosomes have features typical of higher eukaryotes, including large centromeres comprising repetitive sequences, conserved heterochromatin proteins and epigenetic silencing mechanisms, large replication origins, and conserved telomere proteins.
This contrasts with budding yeast, in which a slightly larger genome is divided amongst 16 chromosomes. Budding yeast has simple point centromeres, non-conserved silencing proteins, small origins, and a limited set of telomere proteins. Thus, fission yeast has become a model of choice for eukaryotic chromosome structure.
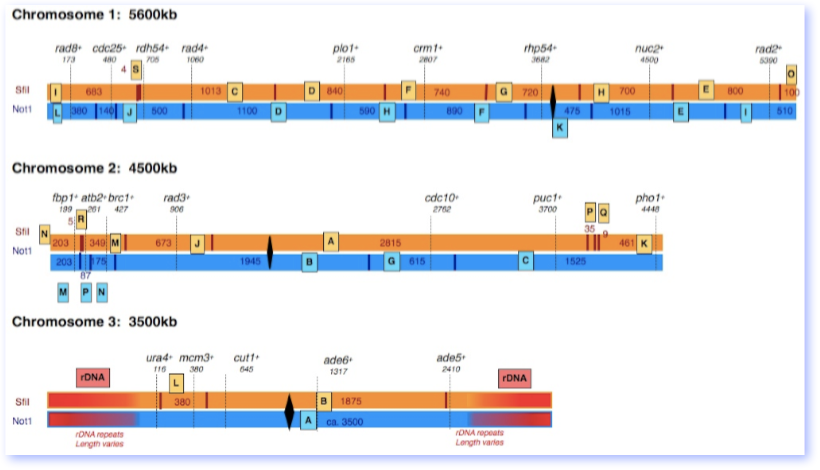
This map of the three pombe chromosomes was derived from Pombase. We’ve included estimated sizes of NotI and SfiI restriction fragments, and indicated marker genes for each fragment. However, you may find that the assignments on a SfiI gel don’t quite match. It’s likely that there is some variation due to genome rearrangements in different regions.
Centromeres
The large fission yeast centromeres are very large, highly repetitive elements, reminiscent of centromeres in multicellular eukaryotes. They all share a central core domain, characterized by incorporation of the centromere-specific histone H3 variant Cenp-A (Cnp1), which forms the basis for the assembly of the kinetochore. Cnp1 in S. pombe as in other organisms is essential for centromere function. Extending out from the central core are two distinct repetitive domains. The inner repeat, or imr, is different in sequence in each centromere. It is a convergent repeat flanking the central core. The outer repeat, or otr, contains two different repetitive elements called dg and dh. These are closely related on all three centromeres, although their orientation and number differs from centromere to centromere.
Mitsuhiro Yanagida writes,
dg is the abbreviation of dogentai, and dh is the next alphabet from g (like amber and ochre), I like to add that Dogentai is the Japanese translation of kinetochore rather than centromere. It literally means ‘moving fundamental body’. In Japanese, centromere is pronounced and written sen-toro-me-a by katakana.
The outer repeats are assembled into heterochromatin, and are defined by binding of Swi6, a fission yeast homologue of Heterochromatin Protein 1. The heterochromatin is important, but not essential for centromere function. It provides structural rigidity that likely helps orient the kinetochore to facilitate association with the correct microtubules. Check out our work on heterochromatin to see a movie of a Swi6 mutant. More about heterochromatin is described below.
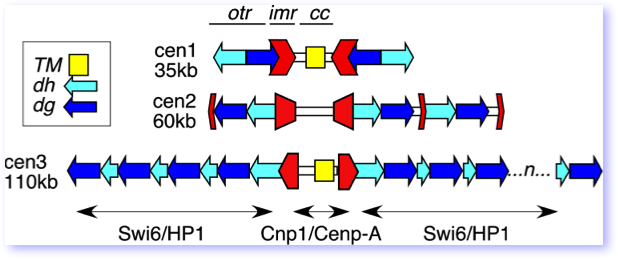
This cartoon of the three S. pombe centromeres shows the relative position of the otr, imr and cc repeats. CenI and CenIII have a common sequence within the central core, TM, that is also found at the mating type locus.
Heterochromatin
Fission yeast is a great model for assembly of silent heterochromatin, as it has kept all the conserved proteins involved in this process. (Many have been lost from budding yeast, which had to figure out a different way to make heterochromatin.) This transcriptionally silent form of chromatin has a characteristic set of epigenetic modifications, most notably methylation of histone H3 at residue K9 (H3K9me). This methylation is performed by the Clr4 methyltransferase, and provides a binding site for the methyl-histone specific chromodomain found in Swi6 (heterochromatin protein 1). The presence of Swi6 “locks down” the chromatin into a transcriptionally inactive state. This is the basic model for the major heterochromatin domains in fission yeast: the centromeres, the telomeres, the mating type loci, and the ribosomal DNA (rDNA).
Naturally, things are somewhat more complicated than this simple description. Mechanisms that establish and maintain the heterochromatin in the different domains have different features, and while Swi6 and Clr4 are common, other proteins are region-specific.
The heterochromatin in the centromere domain is disrupted during each cell cycle, during S phase. This allows a brief window of transcription of non-protein coding sequences. These transcripts are processed by the RNAi protein Dicer into short dsRNA, which is used to target a specialized complex, RITS, back to the site of transcription. RITS contains (amongst others) an RNAi protein Argonaut, and chromodomain protein, Chp1. RITS helps recruit a complex containing Clr4, which methylates histone H3. Chp1 binds to the H3K9me and helps perpetuate the signal. Thus, this brief window of transcription paradoxically leads to re-establishment of silencing. Chp1 is exchanged for Swi6, which also can recruit Clr4 and result in spreading of the heterochromatin until it reaches boundary elements (likely tRNAs) flanking the outer repeats.
Swi6 is also found at the telomere, although RNAi and Chp1 are not involved. There is an overlapping system of silencing that depends on the histone deacetylase Sir2.