 This part of our website tells you about the biology of fission yeast, why we study it, and some aspects of fission yeast biology that we find particular interesting (largely involving cell cycle control, DNA replication and genome integrity questions). Other pages will tell you about our research, and about how to work with S. pombe if you want to use it for your research. We think it’s pretty cool.
This part of our website tells you about the biology of fission yeast, why we study it, and some aspects of fission yeast biology that we find particular interesting (largely involving cell cycle control, DNA replication and genome integrity questions). Other pages will tell you about our research, and about how to work with S. pombe if you want to use it for your research. We think it’s pretty cool.
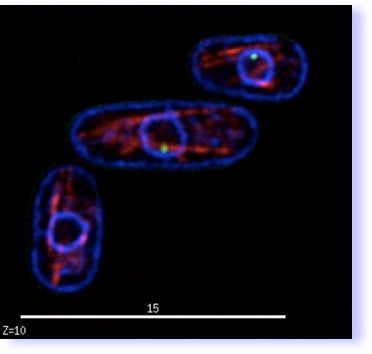
The movie to the right is an optical section through live fission yeast cells that are expressing three fluorescent proteins: one to label the membranes ( blue), one to label the spindle (red) and one to label the centromeres (green). The movie loops through the sections so you can see the organization of these features in the cells.
What’s Yeast?
Yeast is a very general term, and covers a broad range of unicellular fungi that are only vaguely related. You can find out more about various yeasts by visiting the web pages listed in the yeast virtual library. Most people are familiar with brewer’s or baker’s yeast, Saccharomyces cerevisiae, which is a popular laboratory model system as well as a well-domesticated servant. “Yeast infections” typically come from the pathogenic species Candida albicans. However, the species that we study is the fission yeast Schizosaccharomyces pombe, which is a very different organism, not harmful, and easy to grow.
“pombe” means “beer” in Swahili. S. pombe was originally isolated from a millet beer (see the history pages for the original characterization of S. pombe). S. pombe and closely related species have been isolated from grape juice, kambucha tea, and arak. S. pombe particularly useful in the laboratory, where we use it as a model system for cell division.
An aside about beer: Various sources describe pombe beer as millet, maize, or sorghum based. Most sites agree that it is grain based. Millet was used for most beer in the 19th c, other grains today. When I visited Kenya, I was told that traaditional pombe is buried for a period to rot–er, ferment. Other sources suggest that beer-making is traditionally women’s work, and I have a story- quilt from Zimbabwe that illustrates beer making. The taste is described as sour, or even foul.If you use fission yeast to brew with malt and hops (European style), the results are disappointing. My lab has also tried brewing with pombe with unimpressive results. The sulfurous nose was very noticeable and increased with time. But of course, this is not a traditional method, so maybe it tastes better with millet. Or perhaps it’s just an acquired taste. Still, I’ve been told that pombe generates the largest number of foul-tasting compounds for any fermentation and it is typically viewed as a spoilage in winemaking. We have also made bread with it; that was more successful and was actually tasty, although it is not as robust at rising as standard baker’s yeast.
Yeast as a Research Tool
Why is yeast relevant to biomedical research? Normal cell division is central to normal growth and development not only of yeast cells, but of humans as well. But humans are very complicated, so it is easier to establish basic principles in simple model organisms. Importantly, the human versions of a number of yeast cell cycle and DNA repair genes have been found to be directly involved in human cell division. Malfunctions can lead to cancer or birth defects. The normal activity of these genes is most easily studied in the yeast cells, which are readily manipulated in the laboratory. This provides important insights into their mechanisms in humans, which helps direct experiments in more complicated cell types. Studying the control of cell division in yeast is thus very relevant to human health and understanding many clinical disorders. That’s why two researchers studying cell division in yeast shared the 2001 Nobel Prize in Physiology or Medicine, along with a third investigator studying sea urchins! For a description of their landmark research, visit this illustrated presentation from the Nobel site in Sweden.
The real strength of the yeast system is its facile genetics. We can isolate mutants, characterize the genes required for a given process, and move from a relatively abstract genetic description to a very precise, concrete biochemical description of what the gene products actually do. Often, these genes are the same as those found in complex systems, including humans.
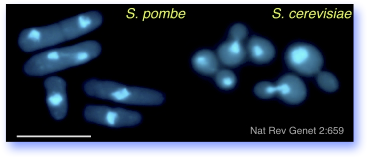
Both S. pombe and Sacch. cerevisiae are harmless, rapidly growing eukaryotes that are popular as model systems to understand basic biological processes. They have many differences, and the comparison between them is uniquely informative, making the yeasts a powerful pair of experimental systems.
For example, many of the genes that regulate normal cell division in complex organisms such as humans also exist and function the same way in yeasts, but each species provides distinct insights. We use fission yeast in particular because recent studies have shown that it organizes its chromosomes very similarly to the organization of human chromosomes.
Phylogeny
Many people ask us how different are S. pombe and Sacch. cerevisiae? The answer is, VERY different. According to some methods of comparison, S. pombe diverged from the ascomycete lineage before Sacch. cerevisiae separated from the filamentous fungi Aspergillus or Penicillium. Based on recent studies, S. pombe is formally classified as an archaeascomycete or taphryinomyces. The NCBI Taxonomy Browser gives the following lineage: Eukaryota/ Fungi/ Ascomycota/ Archiascomycetes/ Schizosaccharomycetales/ Schizosaccharomycetaceae/ Schizosaccharomyces. The phylogeny of S. pombe is a point of ongoing discussion, but an exciting paper on fungal evolution indicates that the divergence between ascomycetes and archaeascomycetes is on the order of 1144 million years ago! That’s over 4 fold longer than previous suspected. This review article describes the lineages of all common model organisms.
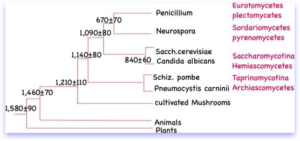
This phylogeny is derived from Nat Rev Genet 2002, 3:838
Not surprisingly, S. pombe biology is quite distinct from that of Sacch. cerevisiae. For a discussion of some of their differences, check out this review.
Recently, several cousins of S. pombe were genome sequenced at the Broad Institute. Schizosaccharomyces japonicus, Schizosaccharomyces octosporalus, and Schizosacharomyces cryophilus are also fission yeasts. Both S. japonicus and S. octosporalus can grow hyphally, making invasive filaments in the agar. They also have 8-spored asci (octads). It will be interesting to see how these species develop as a companion model to S. pombe.
Aside: History
Check out the history section for the history of S. pombe. As for the derivation of our standard lab strain, this information comes from John Armstrong, Paul Young, and Juerg Kohli during a recent conversation on the pombelist mailing list
John notes:
“the standard pombe strains derived from Leupold’s 972 are not connected to Lindner’s original isolate, but apparently originate in some rancid grape juice from Montpelier, Switzerland, in the 1920’s. Some of this is described in the chapter by Munz et al in Molecular Biology of the Fission Yeast. So they have never been to East Africa and never used to make pombe. Secondly, NCYC keep a strain of pombe called NCYC132 which some early workers such as Mitchison and Nurse described. It’s very different from 972 strains and a bit messy to work on. …..The NCYC database describes neither a year of deposition nor a depositor. However I suspect its origins are separate from 972, and they might even be in East Africa.”
However, Juerg Kohli notes (citing Urs Leupold in The Early days of Yeast Genetics book)
“The standard S. pombe strains were isolated by Urs Leupold in 1946 and 1947 from a culture that he obtained from the yeast collection in Delft, The Netherlands. It was deposited there by A. Osterwalder under the name S. pombe var liquefaciens, after he isolated it in 1924 from French wine (most probably rancid) at the Federal Experimental Station of Vini- and Horticulture in WŠdenswil, Switzerland. The culture used by Urs Leupold contained (besides others) cells with the mating types h90 (strain 968), h- (strain 972), and h+ (strain 975).”
Paul Young adds about the mysterious 132::
“At least one difference for 132 is that when challenged with minus nitrogen, 132 does not accelerate mitosis within the largest G2 cohort but rather divides down progressively (Mitchison, pers.com.). As for other isolates I see a recent paper in Pubmed noting that it is common in Kambucha mix and it has also been reported as a (the?) major yeast in cactus beer (pulqa sp?) in Mexico. ”
The next pages in this section of our website include basic overviews of the features of fission yeast biology we find particularly interesting, namely those problems that relate to chromosome maintenance and function. As always, if you find any errors or broken links, please let us know!