We have several projects related to epigenetics and heterochromatin relevant to the broad questions of DNA replication and genome stability. Some background on heterochromatin can be found on the Chromosomes Page.
DNA Replication and the Heterochromatin
As described on the DDK page, we identified an interaction between Swi6 and the DDK kinase. Swi6 is heterochromatin protein 1 (HP1), a conserved protein that binds methylated histones and is important for the establishment of transcriptionally silent heterochromatin (more background on the our chromosomes page). In a collaboration with Robin Allshire, Julie Bailis showed that hsk1 and dfp1 mutants have defective chromosome segregation and centromere cohesin. Hisao Masukata’s lab went on to show that DDK interaction with Swi6 is also important for the early timing of centromere replication.
This movie shows the effect of a swi6 mutation in mutant cells labeled with histone-GFP. Under these conditions, the chromosomes are at risk for inappropriate attachments to the spindle apparatus; some chromosomes may get attached to both poles of the spindle. You can see the cell in the center has this merotelic association and undergoes abnormal segregation and chromosome loss. However, most of the cells manage to divide safely.
Heterochromatin and Genome Stability
See the genome stability page
The Myst family of Histone Acetyltransferases
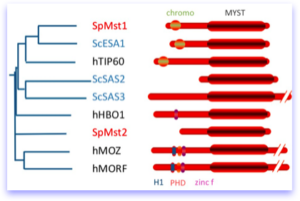 In metazoans, the MCMs have been shown to interact with a protein called Hbo1 which is a member of the MYST family of histone acetyltransferases. This family has numerous members, and is defined mainly by characteristics of the MYST-acetyltransferase domain. The different members of the family have different additional domains, indicated in the figure at right.
In metazoans, the MCMs have been shown to interact with a protein called Hbo1 which is a member of the MYST family of histone acetyltransferases. This family has numerous members, and is defined mainly by characteristics of the MYST-acetyltransferase domain. The different members of the family have different additional domains, indicated in the figure at right.
Although there is no obvious Hbo1 orthologue in fission yeast, there are two members of this family: Mst1, which is the highly conserved Kat5 orthologue corresponding to Tip60/Esa1, and Mst2., which is less well conserved but has features in common with S. cerevisiae HATs called Sas2 and Sas3. Eliana Gómez cloned both mst1+ and mst2+ and performed their initial characterization.
mst1+ is an essential gene. Eliana constructed a temperature sensitive allele and showed that the cells have significant defects in chromosome segregation. Rebecca Nugent followed up on Eliana’s observations. Interestingly, mst1ts mutants have strong genetic interactions with heterochromatin mutants including swi6 and clr4. However, 2-hybrid analysis showed that Mst1 interacts physically with proteins that influence the central core of the centromere, including Msc1and CenpB. Rebecca showed that mst1ts disrupts the kinetochore but not Cnp1 binding in the central core.
Ruben Petreaca has followed up data from Eliana showing that Mst1 interacts with the Rad22 (Rad52) homologous recombination protein. He has dissected the domains of the proteins required for this and assessed the effect on double strand break repair.
mst2+ is not essential. Eliana showed that Mst2 antagonizes the Sir2 histone deacetylase in silencing of the telomere. Working with Anthony Wright’s lab, Rebecca showed that mst2 overlaps with gcn5 in the acetylation of H3K14, and the double mutant is quite damage sensitive. We continue to work on this family of proteins.
Silencing of an ade6+ gene in the telomere in mst2 mutants. In the absence of Mst2, there is increased silencing, and this depends on both Swi6 and Sir2. From Gomez (2004).
Chromosome Passenger Proteins
In collaboration with Tony Hunter’s lab, we also characterized components of the Chromosome Passenger Complex, or CPC. This complex, made up of Ark1, the Aurora kinase, Bir1/Survivin, and Pic1/INCENP, is important for proper attachment of the kinetochore to the centromere in mitosis. Interestingly, we found that a member of the GINS complex, Psf2, interacts with this complex, suggesting that it may be coordinated with the events of S phase.
Reviews
- Forsburg SL (2013) The CINs of the centromere. Biochem Soc Trans. 2013 Dec;41(6):1706-11
- Huberman JA.Subtle interactions between heterochromatin and DNA replication timing. Cell Cycle. 2011 Mar 15;10(6):873-8.
- Bailis, J. M. and Forsburg, S. L. (2004). S phase assembly of centromeric heterochromatin and cohe¬sion. Cell Cycle 3:416-418
- Bailis, J.M. and Forsburg, S.L. (2003). It’s all in the timing: linking S phase to chromatin structure and chromosome dynamics. Cell Cycle 2:303-306
Our Primary research papers on Histones, Heterochromatin, and Chromosome Segregation
- Li PC, Petreaca RC, Jensen A, Yuan JP, Green MD, Forsburg SL. (2013), Replication fork stability is essential for the maintenance of centromere integrity in the absence of heterochromatin. Cell Rep. 2013 Mar 28;3(3):638-45.
- Li PC, Green MD, Forsburg SL. (2013) Mutations Disrupting Histone Methylation Have Different Effects on Replication Timing in S. pombe Centromere. PLoS One. May 1;8(5):e61464. doi: 10.1371/journal.pone.0061464.
- Li, P.C., Chretien, L., Cote, J., Kelly, T.J., and Forsburg. S.L. (2011). S. pombe replication protein Cdc18 (Cdc6) interacts with Swi6 (HP1) heterochromatin protein: region specific effects and replication timing in the centromere. Cell cycle 10(2): p. 323-36.PMC 3025051
- Nugent, R.L., Johnsson, A., Fleharty, B., Gogol, M., Xue-Franzén, Y., Seidel, C., Wright, A.P., Forsburg, S.L. (2010) Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation . BMC Genomics. 2010 Jan 22;11:59. PMC2823694
- Gómez,, E.G., Nugent, R.L., Laría, S., and Forsburg, S.L. (2008) S. pombe histone acetyltransferase Mst1 (KAT5) is an essential protein required for damage response and chromosome segregation Genetics 179(2):757-71. PMC2429872
- Huang, H.-K., Bailis, J.M., Leverson, J.D., Gómez, E.B., Forsburg, S.L. and Hunter, T. (2005) The chromosomal passenger protein Bir1p (Survivin) interacts with Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol. Cell Biol. 25:9000-15. PMC1265766
- Gómez, E.B, Espinosa, J., and Forsburg, S.L. (2005) S. pombe mst2+ encodes a MYST-family histone acetyltransferase that negatively regulates telomere silencing. Mol. Cell Biol. 25:8887-903.. PMC1265769
- Freeman-Cook, L L., Gómez, E.B., Spedale, E.J., Marlett, J., Forsburg, S.L., Pillus, L., and Laurenson, P. (2005) Conserved locus-specific silencing functions of S. pombe sir2+. Genetics169(3):1243-1260. PMC1449530
- Gómez, E.B., Angeles, V.T., and Forsburg, S.L. (2005) A novel screen for S. pombe replication mutants identifies new alleles of cut9+, rad4+, and psf2+. Genetics 169:77-89. PMC1448876
- Bailis, J.M., Bernard, P., Antonelli, R., Allshire, R., and Forsburg, S.L. (2003) Hsk1-Dfp1 is required for heterochromatin mediated cohesion at centromeres. Nat. Cell Biol. 5:1111-1116 (cover).
- Leverson, J.D, Huang, H.-K., Forsburg, S.L. and Hunter, T. (2002) The Schizosaccharomyces pombe Aurora-Related Kinase (Ark1) interacts with the Inner Centromere Protein (INCENP) Pic1, and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell 13:1132-1143. PMC102257