DNA Replication Basics
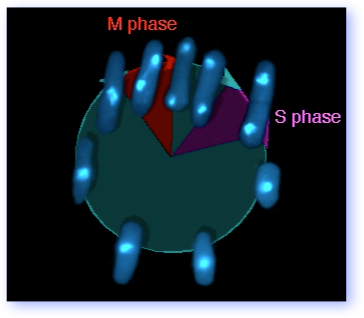
The graphic at the right shows you the fission yeast cell cycle, using photographs of DAPI stained pombe. Although G2 occupies roughly 70% of the cell cycle, the interesting bits to most investigators are DNA replication (S phase), when the chromosomes duplicate, and mitosis (M phase), when the duplicated chromosomes segregate. Although these phases are temporally well separated, events in S phase influence mitosis (for example, by the establishment of sistser chromatid cohesion) and completion of mitosis is linked to re-licensing of the replication origins to allow another round of S phase.
S Phase Entry
Transition into a cycle of division at START requires the product of the p34cdc2 kinase although the targets and cyclin partner(s) of the kinase at this transition point are unknown. This is roughly equivalent to the G0 to G1 transition in mammalian cells. Transition through START also requires the Cdc10p transcriptional activator. This protein offers one way of coupling START to S phase; forming a complex with Res1p and to a lesser extent, Res2p, Cdc10p activates the transcription of several genes known to be required for DNA replication, including cdc22+, encoding the enzyme ribonucleotide reductase; cdc18+, encoding a replication initiator (CDC6 in other systems), and cdt1+. However, Cdc10 has a phenotype that clearly implicates it upstream of these replication proteins (arrested cells are still proficient for mating), and its START-specific targets are unknown.
Replication Origin Assembly
The earliest stages of S phase involve the assembly and activation of the individual replication origins–there are about 400 of them in fission yeast, and they have been mapped using genome-wide arrays and protein binding assays. Fission yeast origins are generally A/T rich sequences of ca. 1kb. They do not have a consensus sequence (unlike budding yeast) so they are defined functionally. There is considerable stochastic variability in whether an origin fires or not. Some are efficiently used in every cell cycle; others are less likely to fire, and may provide a “backup function”.
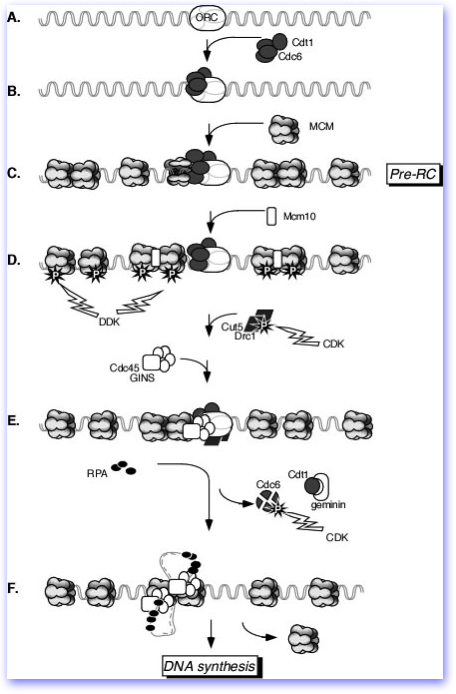
Origins are prepared for use by assembly of a pre-replication complex, or preRC, which actually forms during M phase. This occurs by the sequential loading of Cdc18(aka Cdc6) and Cdt1 , and the 6 members of the MCM complex: Mcm2,Mcm3, Mcm4, Mcm5, Mcm6, and Mcm7. This only occurs individual replication origins that are already bound by the Origin Recognition Complex, ORC. The MCM complex is the primary focus of our lab and you can learn more about our work and the MCMs starting here.
The cartoon at right (from Forsburg, MMBR 2004) diagrams the steps in replication origin assembly and activation; however, the precise order of some of these steps with lesser-known components remains unclear.
One puzzle about the MCM complex is that it is strikingly abundant, vastly exceeding the number of origins in the cells. This is known as the MCM paradox. MCM proteins liberally decorate unreplicated chromatin, suggesting that they may mark it it some way as replication competent, or licensed. For this and other reasons, they were historically identified as licensing factor.
Once these proteins are assembled, the origin is poised to fire. Without these proteins, the origin is inert. Therefore, origin firing is accompanied by origin inactivation, primarily through the regulated destruction of the Cdc18 protein which ensures that in each cell cycle, an origin fires once and only once.
What is clear is that the cyclin dependent kinase Cdc2 (aka CDK1) has a crucial role in controlling origin assembly and utilization. Pre-RC proteins assemble on the origin when CDK is low. When CDK is high, the origin fires and Cdc18 is destroyed. Thus the CDK oscillator also controls origin assembly.
Initiation
Activation of the replication origins is accomplished by two kinases: CDK, the central cell cycle oscillator, and DDK, the Dbf4-dependent kinase. DDK consists of a catalytic subunit (Hsk1 in S. pombe, Cdc7 in other systems) and a regulatory subunit (Dfp1 in S. pombe, Dbf4 elsewhere). Where CDK affects global regulators, DDK appears to act locally at each origin, by influencing the activity of the MCM complex. Activation by DDK converts the MCM complex from an assembly factor to a helicase essential for elongation of DNA synthesis. Additional proteins are loaded, including Sld3, the GINS complex (Psf1, Psf2, Psf3, and Sld5), and Cdc45. Evidence suggests that these are processivity factors for the MCM helicase, and Cdc45 is rate limiting for replication.
The initial steps of unwinding by the MCM helicase expose short single stranded regions of DNA. Following this, there is the recruitment of DNA polymerases and primase for leading and lagging strand synthesis. The replisome that includes the MCMs is also linked to chromatin remodelers (the FACT complex) and proteins involved in establishing sister chromosome cohesion. Additionally, replication fork protection proteins, including Mrc1, Swi1, and Swi3, contribute to fork efficiency and maintenance.
The MCM helicase is also a contributor to replication fork stability and a key player in maintaining the fork during stops and starts.
Our Research
We work on a variety of aspects of MCM function and DNA replication, both in mitosis and meiosis. Go to the research page to learn more about our work.
Replication in live cells
In this movie, watch the dynamics of PCNA-GFP in live cells. The intensity of PCNA will change with DNA content and can be used as a metric for S phase.