This page describes our research on MCM proteins (continued on interior pages). For background information, check out the fission yeast pages on pombe, the cell cycle, DNA replication, and meiosis.
The MCM Proteins
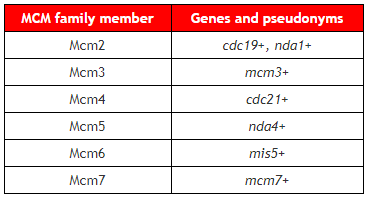
Much of our work derives from our longstanding study of the fission yeast MCM proteins. “MCM” stands for “MiniChromsome Maintenance defect”, which is the phenotype used to isolate the founding members of this family in budding yeast in a rich screen performed years ago by Bik Tye. The fission yeast mcm genes were identified genetically: either as cdc mutants with defects in the the Cell Division Cycle, nda mutants with a phenotype of nuclear division arrest, or mis mutants with chromosome mis-segregation defects. Classic molecular cloning of these genes allowed a number of labs to identify them as members of the conserved MCM family and they are now known by those names. Our lab cloned cdc19+ (mcm2+), mcm3+, and mcm7+ .
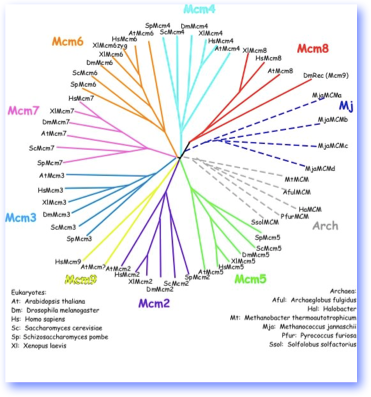
The MCMs are a family of proteins that are a sub-type of the huge AAA+ ATPase family. They are an ancient family, with some relatives found in the Archaea. All MCMs are related to one another, and share distinct features particularly in their ATPase domain. Outside of this region, the MCMs are distinct from one another, so that (for example) Mcm2 from S. pombe is more closely related to Mcm2 from S. cerevisiae, than to S. pombe Mcm3. All eukaryotes have at least six MCMs which are known as Mcm2- Mcm7. There are also Mcm8 and Mcm9 in some metazoans, but their contribution to MCM function is still somewhat obscure.
The figure shows a tree including representatives of the MCM family from eukaryotes and from Archaea, color coded by family. If you click on it, you will see a bigger version (copyright SLF). Note there is a zygotic form of Mcm6 in Xenopus. There’s a little confusion in the nomenclature of Mcm8 and Mcm9, which are found only in multicellular organisms, but sequence comparisons can be used to classify them.
 Aside: What happened to Mcm1? The original MCM screen by Tye’s lab isolated mutants that fell broadly into two classes: replication mutants and chromosome segregation mutants. Either defect will lead to minichromosome loss, and the screen was very successful, yielding 21 “mcm” mutants. However, only upon cloning and sequencing was it determined that several of these fall into a single protein family. That family became known as Mcm2 – Mcm7. There is an Mcm1, but it is a transcription factor involved in cell cycle specific regulation, and is NOT a member of the MCM family.
Aside: What happened to Mcm1? The original MCM screen by Tye’s lab isolated mutants that fell broadly into two classes: replication mutants and chromosome segregation mutants. Either defect will lead to minichromosome loss, and the screen was very successful, yielding 21 “mcm” mutants. However, only upon cloning and sequencing was it determined that several of these fall into a single protein family. That family became known as Mcm2 – Mcm7. There is an Mcm1, but it is a transcription factor involved in cell cycle specific regulation, and is NOT a member of the MCM family.
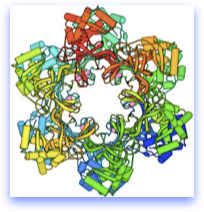
The structure at top is the archael MCM homohexamer, solved by our colleague Xiaojiang Chen’s lab.
The MCM Complex
The 6 MCM proteins form a ring-shaped complex, in a characteristic order. Each ATP-binding site has contributions from two adjacent subunits: the Walker A-B ATPase motifs of one partner and the “arginine finger” of the other. Although the right looks symmetrical, biochemical analysis from several groups has shown that that only three of the sites are active as an ATPase. THus, as Eliana Gómez showed, some of the MCM subunits actually tolerate mutations in the ATPase domain without killing the cell. However, there is no genetic evidence that any of the MCMs function separately; their mutant phenotypes in yeast are essentially identical.
The ring breaks up into characteristic parts, which Dan Sherman in our lab characterized. The Mcm3-5 pair fall of the complex rather easily. Mcm4-6-7 form a tightly bound “core” to which Mcm2 binds. The connection between Mcm2 and Mcm5 appears so fragile that it is likely to be the gate by which the MCMs are installed on the DNA. In fact, it’s tempting to speculate that the MCM complex may also be a filament, not just a ring.
There are clearly alternative MCM complexes. In metazoa, these may include Mcm8 and Mcm9. There’s also an alternative complex that replaces Mcm2 with a non-Mcm protein, MCM binding protein 1, aka MCM-BP or in pombe, Mcb1, worked on by Lin Ding. This is an abundant protein that may antagonize the MCM complex, although the details are still uncertain.
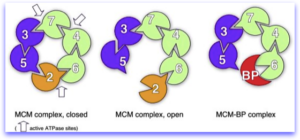 MCM complexes. Left, the MCM complex on chromatin hydrolyzes ATP at three of the six possible sites. middle, Mcm2 acts as a gating subunit. Right, in some cases, Mcm2 may be replaced by MCM-BP, which renders the complex inactive. This might be a way to remove the complex from the chromatin.
MCM complexes. Left, the MCM complex on chromatin hydrolyzes ATP at three of the six possible sites. middle, Mcm2 acts as a gating subunit. Right, in some cases, Mcm2 may be replaced by MCM-BP, which renders the complex inactive. This might be a way to remove the complex from the chromatin.
MCMS in the Nucleus and on Chromatin
In most eukaryotes, MCMs are found in the nucleus all the time (the exception is budding yeast, where they go in and out of the nucleus in a cell-cycle dependent fashion fashion). Sally Pasion showed that Mcm2 and Mcm3 provide the nuclear targeting information to the complex; Mcm2 may be responsible for the localization of Mcm4-7, while Mcm3 brings in Mcm5. If the complex in the nucleus is disrupted, the excess subunits are exported. Thus, there is a role for the nucleus is regulation of MCMs, even though it is not cell cycle dependent. This way, the cell makes sure that MCMs are stoichiometric and there are no rogue subunits in the nucleus.
Early work in mammals showed that MCMs liberally decorate unreplicated chromatin, which was why they were first identified as “licensing factor”. As replication proceeds, these excess MCMs are removed from the chromatin, so some have suggested that they mark the unreplicated part of the genome (or license it) for replication. However, genome wide localization of the MCMs suggests they are specific to the origin. Thus, the “remote” MCMs are not bound at specific sites, and it’s attractive to speculate that they have a different relationship to the chromatin. It’s interesting to speculate that this may have something to do with the ability of Mcm2 to bind histone. We suggest there are three distinct pools of MCM complexes in the nucleus:
- “active MCM” at the origin/fork, linked to the DNA and functioning as a helicase
- “remote MCM” decorating unreplicated chromatin
- “soluble MCM” not associated with the chromatin at all.
Activation of the MCMS
The transition from origin assembly factor to replicative helicase occurs in response to the DDK and CDK kinases, which results in loading of some additional proteins, including the Sld3, Cdc45 and the GINS complex. It is likely that these are processivity factors that make the MCM helicase proficient and processive. Several additional players include Rad4/Cut5, a protein with multiple functions in checkpoint and repair (investigated by Rania Siam) and Mcm10/Cdc23, which may link the complex to DNA polymerases. It appears that there is relatively little MCM required to “get going” in replication; most mcm mutants synthesize a substantial amount of DNA even at the restrictive temperature. It takes considerable effort to deplete MCMs enough to arrest the cells prior to DNA replication.
The Conundrum: Many MCM Mutations don’t block DNA Replication
All this is very pretty, but there’s a puzzle. The majority of mcm mutants cause cells to arrest with a 2C (ie, apparently replicated) DNA content. This includes most temperature sensitive alleles, as well as germinating spores containing a disruption in an mcm gene. This is accompanied by DNA damage and activation of the DNA damage checkpoint (see our genome stability page for more). But how can this be, if the MCMs are essential for initiation?
The answer is apparently that the amount of MCM required for initiation is quite small, but quite a lot is needed for completion of S phase. The spores with the disruption package enough from the parental cell to allow initiation and DNA synthesis. Only an mcm4ts-degron allele, constructed by Stephen Kearsey’s group, is sufficient to block initiation at a high temperature (although it remarkably does not block nuclear division; see our genome stability page for more). Thus, most of our mcm-ts alleles are separation of function alleles, which will become important as we discuss meiosis.
-
There are a number of questions about MCMs that still haven’t been answered in a satisfactory way.
- The original genetic analysis of mcm mutants showed cells arrested with a 2C (replicated) content of DNA. But why, if they have completed replication, are they still unable to proceed? Our data (next page) suggest this reflects DNA damage and fork collapse. Additionally, there may be regions of the genome that are refractory to replication without a full complement of MCMs.
- Why are there so MANY MCMs? Some estimates suggest ca. 40,000 MCMs in a yeast cell, with only 400 origins. This excess of MCM is called the MCM paradox.
- How does the cell keep the helicase linked to the replisome? If the MCMs “get away” from the replisome, that could lead to excessive unwinding and DNA instability. Candidate proteins include Mrc1 and the Fork Protection Complex (Swi1 and Swi3) on the leading strand, and Mcl1/Ctf4 on the lagging strand.
- What about other MCM functions? There are suggestions that MCMs affect transcription and recombination.
- There’s also evidence that MCMs localize to the centrosomes. What’s up with that? perhaps it integrates this organelle with the replication cycle.
-
- Brewster AS, Chen XS. Insights into the MCM functional mechanism: lessons learned from the archaeal MCM complex. Crit Rev Biochem Mol Biol. 2010 Jun;45(3):243-56. Review. PubMed PMID: 20441442; PubMed Central PMCID: PMC2953368.
- Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009 Dec;73(4):652-83. Review.
- PubMed PMID: 19946136; PubMed Central PMCID: PMC2786579.
- Bailis JM, Forsburg SL. MCM proteins: DNA damage, mutagenesis and repair. Curr Opin Genet Dev. 2004 Feb;14(1):17-21. Review. PubMed PMID: 15108800.
- Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004 Mar;68(1):109-31. Review. PubMed PMID: 15007098; PubMed Central PMCID: PMC362110.
.
-
- Sabatinos, S.A., Ranatunga, N.S., Yuan, J.-P., Green, M.D., and Forsburg, S.L. (2015) Replication stress in early S phase generates apparent micronuclei and chromosome rearrangement in fission yeast. Mol Biol Cell. 2015 Aug 5. pii: mbc.E15-05-0318. . [Epub ahead of print]
- Slaymaker IM, Fu Y, Toso DB, Ranatunga N, Brewster A, Forsburg SL, Zhou ZH, Chen XS. (2013) Nucleic Acids Res. 2013 Mar 1;41(5):3446-56.
- Ding, L. and Forsburg, S.L. (2011) Schizosaccharomyces pombe MCM binding protein (MCM-BP) antagonizes MCM helicase. J. Biol. Chem. in press doi:10.1074/jbc.M111.282541
- Singh SK, Sabatinos S, Forsburg S, Bastia D. (2010) Regulation of replication termination by Reb1 protein-mediated action at a distance. Cell 142(6):868-78.
- Bailis, J.M., Luche, D.D. Hunter, T., and Forsburg, S.L. (2008) MCM proteins interact with checkpoint and recombination proteins to promote S phase genome stability Mol. Cell. Biol. 28:1724-38. PMC2258774
- Siam, R., Gómez, E.B., and Forsburg, S.L. (2007) S.pombe Rad4/Cut5 protein modification and chromatin association changes in DNA damage. DNA Cell Biol. 26:565-75.
- Huang, H.-K., Bailis, J.M., Leverson, J.D., Gómez, E.B., Forsburg, S.L. and Hunter, T. (2005) The chromosomal passenger protein Bir1p (Survivin) interacts with Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol. Cell Biol. 25:9000-15. PMC1265766
- Gómez, E.B., Angeles, V.T., and Forsburg, S.L. (2005) A novel screen for S. pombe replication mutants identifies new alleles of cut9+, rad4+, and psf2+. Genetics 169:77-89. PMC1448876
- Dolan, W.P., Sherman, D.A., and Forsburg, S.L. (2004) S. pombe replication protein Cdc45/Sna41 requires Hsk1/Cdc7 and Rad4/Cut5 for chromatin binding. Chromosoma 113:145-156
- Gómez, E.B., Catlett, M.G., and Forsburg, S.L. (2002) Different in vivo phenotypes associated with ATPase motif mutations of S. pombe MCM proteins. Genetics, 160:1305-1318. PMC1462049
- Liang, D.T., and Forsburg, S.L. (2001). Characterization of S. pombe mcm7+ and cdc23+ (MCM10) and interactions with replication checkpoints Genetics 159: 471-486. PMC1461838
- Forsburg, S.L. and Hodson, J.A. (2000) Mitotic replication initiation proteins are not required for S. pombe pre-meiotic S phase.Nat. Genet. 25:263-268
- Pasion, S.G. and Forsburg, S.L. (1999) Nuclear localization of fission yeast Mcm2/Cdc19p requires MCM complex formation. MolBiol. Cell 10: 4043-4057. PMC25742
- Snaith, H.A. and Forsburg, S.L. (1999) Re-replication in fission yeast requires MCM proteins and other S phase genes. Genetics 152: 839-851. PMC1460649
- Liang, D.T., Hodson, J.A. and Forsburg, S.L. (1999) Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J. Cell Sci. 112:559-567
- Sherman, D. A. and Forsburg, S. L. (1998). S. pombe Mcm3p, an essential nuclear protein, associates tightly with Nda4p (Mcm5p).Nucl. Acids Res. 26:3955-3961. PMC147805
- Sherman, D.A., Pasion, S.G., and Forsburg, S.L. (1998) Multiple domains of fission yeast Cdc19p (MCM2) are required for association with the core MCM complex. Mol. Biol. Cell 9:1833-1845. PMC25423
- Gould, K.L. , Burns, C.G., Feoktistova, A., Hu, C.-P., Pasion, S.G., and Forsburg, S.L. (1998). Fission yeast cdc24+ encodes a novel replication factor required for chromosome integrity. Genetics 149:1221-1233. PMC1460225
- Forsburg, S.L., Sherman, D.A., Ottilie, S., Yasuda, J.R., and Hodson, J.A. (1997). Mutational analysis of the S. pombe Cdc19p DNA replication protein. Genetics 147: 1025-1041. PMC1208231
- Forsburg, S.L. and Nurse, P. (1994). The fission yeast cdc19+ gene encodes a member of the MCM family of replication proteins. J. Cell Sci. 107:2779-2788
- Kelly, T.J., Martin, G.S., Forsburg, S.L., Stephen, R.J., Russo, A., and Nurse, P. (1993). The fission yeast cdc18+ gene product couples S-phase to START and mitosis. Cell 74: 371-382.