Much of our interest of late has been in determining how the panel of molecules in which we are interested, primarily but not only MCMs, contribute to overall genome stability (not just DNA synthesis). A substantial part of this investigation has been based on sophisticated imaging methods on both live and fixed cells.
MCMS are required for Fork Stability
Years ago, Debbie Liang (a student in the lab) made two important findings. First, she showed that mcm-ts alleles had significant problems even at permissive temperatures, such that reduced dosage of a single MCM protein causes increased rates of chromosome loss and recombination (1999). Second, she showed that the mcm mutants are irreversible: they don’t recover if you shift them back down to permissive termperature after an arrest at 36°C, even though they appear to have a 2C DNA content. Thus, the problem isn’t in bulk DNA accumulation.
Debbie suggested this was due to DNA damage, because if she deleted the Chk1 damage checkpoint, the cells continued into a lethal mitosis. This can be seen in the phenotypes of the cells as seen on the left below. This was confirmed by Julie Bailis (2008), who showed that mcm mutants accumulate markers of DNA damage, including phosphorylation of histone H2A, and numerous foci of the homologous recombination protein Rhp51. Fast forward to 2015, and Sarah Sabatinos determined that this arrest depends on the Mus81 endonuclease. That is, something about the activation of Mus81 leads to activation of the damage checkpoint. Without Mus81, mcm4-ts mutants fail to arrest the cell cycle.
The damage markers that accumulate in mcm mutants are superficially similar to the markers that accumulate under conditions known to cause replication fork collapse. Normally, the Cds1 checkpoint kinase (Chk2, Rad53 in other systems) is responsible for maintaining the fork for example when cells are starved by nucleotides (treated with the drug hydroxyurea). Cds1 has numerous targets in this process including the DDK kinase(which prevents additional origin firing), the cell cycle machinery, and the endonuclease Mus81.
Julie Bailis found that if she arrested cells in HU, allowing forks to stall, and then inactivated the MCMs (in the continuing presence of HU), that was sufficient to induce damage detected by H2A phosphorylation. Julie also showed that one of our mcm4ts alleles was rescued by pre-treatment with HU. Under those conditions, Cds1 is activated, and Doug Luche showed evidence for Mcm4 being phosphorylated in a Cds1 manner in HU, suggesting that MCMs are a proximal target of Cds1 in the maintenance of the replication fork.
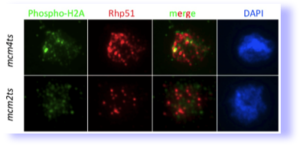 mcm mutants cause DNA damage (from Bailis et al, 2008). Spread nuclei were stained for markers phosphorylated histone H2A, a marker for double strand breaks. They also accumulate extensive foci of the homologous recombination protein Rhp51 which recognizes a wide range of lesions.
mcm mutants cause DNA damage (from Bailis et al, 2008). Spread nuclei were stained for markers phosphorylated histone H2A, a marker for double strand breaks. They also accumulate extensive foci of the homologous recombination protein Rhp51 which recognizes a wide range of lesions.
![]() Chromatin fiber spreading. Proteinated chromatin fibers are spread from nuclei and analyzed by immunofluorescence for Mcm2, BrdU(labeling new DNA synthesis), and total DNA.
Chromatin fiber spreading. Proteinated chromatin fibers are spread from nuclei and analyzed by immunofluorescence for Mcm2, BrdU(labeling new DNA synthesis), and total DNA.
The Architecture of Collapse, The Strategy of Recovery
The work on MCMs led Sarah Sabatinos to ask what REALLY happens during fork collapse? And, even more interesting, what happens during recovery–how are forks restarted? To investigate the dynamics of this process, Sarah developed a unique combination of imaging methods, including single cell analysis of live cells treated with, and then released from HU , labeling of new DNA synthesis with thymidine analogues, and analysis of chromatin fibers for a visual architecture of the replication fork (see above) This work has led to some fascinating insights including distinctly different phenotypes in different mutants that by the old “snapshot” methods of analysis appeared indistinguishable. These methods can now be applied broadly to a wide spectrum of replication mutants to distinguish their defects from one another.
In an initial study (Sabatinos 2012), she showed that cds1∆mutants treated with hydroxyurea actually do not arrest DNA synthesis. This was determined looking at accumulation of EdU, rather than flow cytometry. Why had we we all missed this? Turns out, the cds1∆ cells are loaded with single strand DNA, and that disrupts the accurate quantitation of DNA by FACS.
Next, Sarah examined both RPA and Rad52 foci and found that the signals vary considerably in different forms of replication stress (Sabatinos 2012, 2015). The cds1∆ +HU cells accumulate so much ssDNA (RPA) that the signal is bright and pan-nuclear. The original mcm4-ts allele accumulates dispersed puncta, consistent with late fork failure at distant places (perhaps fragile sites?) while the mcm4-dg allele (described below). accumulates a single large focus that we call a mega-focus. Thus, although we would argue that all of these suffer some form of Fork Collapse, the actual structures are distinct.
cds1 cells expressing fluorescently tagged RPA (false-colored yellow) and Rad52 (false-colored magenta) during HU treatment and following release. There is significant accumulation of RPA during release. A fraction of cells continues to divide.
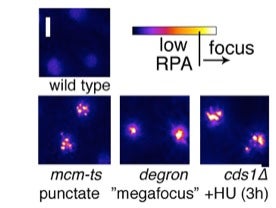 RPA focus patterns during replication collapse are different in each mutant, but rarely develop in wild type at 36°C . Multiple (3+) punctate RPA foci form in mcm-ts nuclei after 4h 36°C and later become a pan-nuclear RPA signal like that observed in cds1∆+HU . A unique “megafocus” of bright, compact RPA forms in mcm4-degron . Heat map scale (top) and 2µm scale are shown.
RPA focus patterns during replication collapse are different in each mutant, but rarely develop in wild type at 36°C . Multiple (3+) punctate RPA foci form in mcm-ts nuclei after 4h 36°C and later become a pan-nuclear RPA signal like that observed in cds1∆+HU . A unique “megafocus” of bright, compact RPA forms in mcm4-degron . Heat map scale (top) and 2µm scale are shown.
 Following release from restrictive temperature, mcm4-dg generates ultra fine anaphase bridges labeled with RPA-CFP
Following release from restrictive temperature, mcm4-dg generates ultra fine anaphase bridges labeled with RPA-CFP
Abnormal Divisions without Replication
Recently, we found that the mcm4-dg mutation, originally isolated by Stephen Kearsey, blocks DNA synthesis very early following initiation, yet does not activate the checkpoint. The cells undergo a highly unusual, unbalanced segregation that includes apparent micronuclei and ultra fine anaphase bridging, accompanied by genome rearrangement. This leads us to propose that the biggest defect in replication may not be the absence of DNA synthesis but the effort to undergo mitosis with a hopelessly entangled genome.
The Pericentromere is a Fragile Site
Genetic interactions between swi6 (the Heterochromatin-Protein 1/HP1 orthologue) and mutants that affect genome stability such as rhp51 or mus81 led us to investigate how the mechanisms that we study during DNA replication are affected by heterochromatin. Pao-Chen Li, assisted by Ruben Petreaca and Amanda Jensen, showed that there is a synthetic phenotype between heterochromatin mutants and mutations affecting replication fork stability. The combination of both these mutations leads to highly unstable outer repeats and a dramatic increase in rearrangement and recombination. Thus, fork stability is particularly important in repetitive sequences that are unprotected by heterochromatin. During the course of this work, we isolated several strains with interesting chromosome rearrangements which we continue to study.
Investigating Nucleases and Helicases
What other proteins contribute to maintenance of genome stability, outside the core replisome? Lin Ding examine Rad8 (ScRad5, human HLTF), which has both helicase and ubiquitin ligase domains. Surprisingly, and in contrast to other systems, she found no striking phenotype associated with the loss of the helicase domain. Rather, it appears the most important function for Rad8 is its ubiquitin ligase. However, when normal homologous recombination is disrupted, there is a minor effect of the helicase mutation.
Tara Mastro found that Rad16 (Swi9, the XPF/Rad1 orthologue) has evidence for genome instability and particularly chromosome mis-segregation. This effect is most dramatic in meiosis. Her data indicate that Rad16 has functions beyond its well-studied role in nucleotide excision repair.
Reviews on Genome Stability
- Sabatinos, S.A. and Forsburg, S.L. (2013) Preserving the Replication Fork in Response to Nucleotide Starvation: Evading the Replication Fork Collapse Point. In: The Mechanisms of DNA Replication, Dr. David Stuart (Ed.), ISBN: 978-953-51-0991-4, InTech, DOI: 10.5772/51393. Open Access
- Forsburg, S.L. (2008) The MCM helicase: at the interface of checkpoints and the replication fork. Bioch. Society Trans. 36:114-9.
- Forsburg, S.L. (2004). Eukaryotic MCM proteins: beyond replication initiation. Mol. Micro. Biol. Rev. 68:109-131 .. PMC362110
- Bailis, J.M. and Forsburg, S.L. (2004). MCM proteins: DNA damage, mutagenesis, and repair. Curr. Op. Gen. Dev. 14 :17-21
- Sabatinos, S. A. (2010) Replication Fork Stalling and the Fork Protection Complex. Nature Education 3(9):40
- Sabatinos, S. A. (2010) Recovering a Stalled Replication Fork. Nature Education 3(9):31
Our Primary Research Publications on Genome Stability
- Sabatinos, S.A., Ranatunga, N.S., Yuan, J.-P., Green, M.D., and Forsburg, S.L. (2015) Replication stress in early S phase generates apparent micronuclei and chromosome rearrangement in fission yeast. Mol Biol Cell. 2015 Aug 5. pii: mbc.E15-05-0318. . [Epub ahead of print]
- Mastro TL, Forsburg SL. (2014) Increased Meiotic Crossovers and Reduced Genome Stability in Absence of Schizosaccharomyces pombe Rad16 (XPF). Genetics. Oct 6. pii: genetics.114.171355. [Epub ahead of print]
- Ding L, Forsburg SL. (2014) Essential domains of Schizosaccharomyces pombe Rad8 required for DNA damage response. G3 (Bethesda). May 28;4(8):1373-84. doi: 10.1534/g3.114.011346.
- Li PC, Petreaca RC, Jensen A, Yuan JP, Green MD, Forsburg SL. (2013), Replication fork stability is essential for the maintenance of centromere integrity in the absence of heterochromatin. Cell Rep. 2013 Mar 28;3(3):638-45.
- Sabatinos, S.A., Mastro, T.L., Green, M.D., and Forsburg, S.L. (2013) Nucleoside analogues create DNA damage and drug sensitivity in fission yeast Genetics. 2013 Jan;193(1):143-57.
- Sabatinos, S.A., Green, M.D., and Forsburg, S.L. (2012) Continued DNA synthesis in replication checkpoint mutants leads to fork collapse. Mol Cell Biol. 2012 Dec;32(24):4986-97.
- Singh SK, Sabatinos S, Forsburg S, Bastia D. (2010) Regulation of replication termination by Reb1 protein-mediated action at a distance. Cell 142(6):868-78.
- Bailis, J.M., Luche, D.D. Hunter, T., and Forsburg, S.L. (2008) MCM proteins interact with checkpoint and recombination proteins to promote S phase genome stability Mol. Cell. Biol. 28:1724-38. PMC2258774
- Hodson, J.A., Bailis, J.M. and Forsburg, S.L. (2003) Efficient labeling of fission yeast S. pombe by thymidine and BUdR. Nucl. Acids Res. 31:e134. PMC275491
- Liang, D.T., Hodson, J.A. and Forsburg, S.L. (1999) Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J. Cell Sci. 112:559-567