One of the most common questions we get is “why won’t my diploid behave?” This page provides information on the nuts’n’bolts of genetic analysis: mating types and crosses, making diploids, working with h90 strains, and analysing meiotic products by random spore analysis or by tetrad dissection. Stable, non-sporulating diploids can also be constructed for specific uses. You can use diploids for plasmid complementation experiments of essential genes.
Mating Types
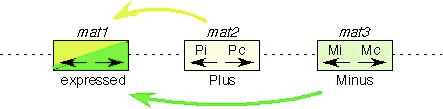
The mating type locus mat1 of S. pombe has three possible alleles: h+ (plus), h- (minus), and h90 (these should normally be written h+,h-, and h90, but that’s harder to read on the web). h- and h+ are heterothallic strains–that means you have to provide them a partner of the opposite mating type if you want them to mate. The h90 allele makes a homothallic strain–that means that it is capable of mating with itself. h90 really means that the cells are constantly switching mating types between h+ and h-, so that any colony actually contains a mixture of h- or h+ mating types, and therefore there is always a partner available. The 90 indicates that 90% of the cells can switch. The “reserved” information is stored in the silent mating loci, mat2 for the plus, or P information, and mat3 for the minus, or M information. The mechanism of switching is quite different from that in S. cerevisise.
Cells expressing the M allele, are h-, and produce M factor. h+ cells express the P allele , and produce P factor. Both M and P factor are required in the same cell for meiosis, which thus limits sporulation to diploids. Unlike Saccharomyces, however, haploid mating functions are not repressed in pombe diploids, so that on occassion diploids will mate with each other.
Crosses & ASCI
Most of the time, you don’t need to select diploids to do a simple cross–just set up a mating mixture of two haploid strains on malt extract and wait 2-3 days. The mating cells will form transient diploids (zygotes) and then proceed directly into meiosis. The plate can be kept in the fridge for a few days after spores form. Note that fission yeast asci fall apart on their own (no glusalase required) so if they go too long, they fall out of their tetrads.
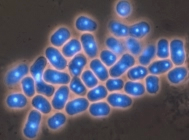
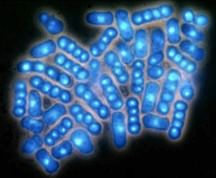
Asci produced from direct mating and meiosis are typically curved in a zig-zag or a banana shape (see h90 picture). These are called zygotic asci, and they reflect the shape of the mated pair of cells, or zygote. However, if a diploid is created and then sporulated, the ascus looks like a single cell and is linear (see diploid picture). These are called azygotic asci. The spores look more tightly packed. An occassional zygotic ascus in a diploid strain usually represents two diploids mating.
h90 strain (zygotic asci)
h+/h- diploid (azygotic asci)
Working with H90 Strains
The h90 marker can be used as either h- or h+ in crosses (since half of the population at any given time is h-, and half is h+). That is, you can cross your h90 strain with an h+ or h- partner. But the h90 colony is likely to provide its own partner rather than use the one you give it, simply because of proximity. Thus, the best way to cross it is to make a diploid first, using complementing markers; e.g., h90 ade6-m210 X h+ ade6-M216 (see diploid section for more detail; any pair of complementing markers can be used). Upon subsequent sporulation, the h90 allele will segregate in normal Mendellian fashion, giving a 2:2 ratio with the h+ allele. The same sort of cross can be done with h-.
If you are used to working with cerevisiae, you may think h90 is similar to using an HO strain. However, in pombe, the mutations in mating type are not in the switching apparatus (as in an ho mutant, which lacks an endonuclease), but in the mating type locus itself. That is, h+ and h- are still potentially switchable, they just don’t have the correct silent information to switch into place. Most h+ strains have rearranged the mating type loci so that they switch plus to plus; these can revert to h90 at about 1 in 103. In contrast, most h- strains have lost all P information and are constitutively minus.
Also, unlike cerevisiae, pombe only mates if it is starved, and the diploid generally is unstable and sporulates immediately. Thus unless you make an effort to keep cells diploid, for example by complementing markers, they will be haploid, regardless of their mating type!
Working with Diploids
Unlike budding yeast, fission yeast does not want to be diploid. Normally, mating is followed immediately by meiosis. (This is why fission yeast investigators do linkage analysis more than complementation testing.) However you can recover diploids and force them to grow vegetatively in the lab. To do this, you must remove cells from the mating mixture on malt extract, return them to normal media, and select for complementing markers. We take cells off of ME after 12 and 24 hours of mating and streak onto selective EMM. Charlie Hoffman recommends selecting diploids (where possible) at 36°C, since high temperatures will repress meiosis. Obviously you need to be sure that any temperature sensitive markers are heterozygous, and fully recessive before you try this!
The most popular set of markers is ade6-M210 and ade6-M216, which complement intragenically so that only the diploid is Ade+. Of course any complementing markers can be used (although watch out for wild type recombinant haploids.) Note that diploid cells are both longer and wider than haploids; it doesn’t take long to learn to recognize them under the microscope.
Because diploids are partially induced for meiosis, they are prone to sporulate even on EMM (minimal media). Your diploid colonies on your EMM plates have probably sporulated in the middle of the colony. Therefore, it is useful to restreak diploid strains onto rich YES media with phloxin B, which stains diploid colonies a darker pink. YES will suppress sporulation more efficiently than EMM, but even so, there is a tendency to accumulate sporulation-deficient cells in a diploid strain that is maintained too long, so the best thing to do is to make it fresh where possible, keep it on YES, and freeze it down quickly if you need to keep it. Diploids should be checked frequently for the ability to sporulate by replica plating a streak of single colonies to malt extract (mating) media, and iodine staining to identify those still sporulation-competent.
How to make a Diploid
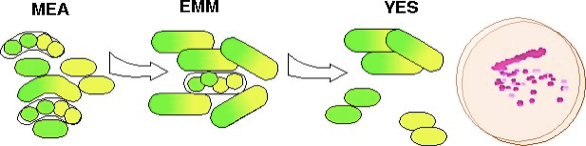
This diagram shows a flow chart of making a diploid. First, you mate them on MEA or SPAS. Your mating mixture will include haploids, diploids, and zygotic asci that have completed sporulation. Second, you will select for the complementing markers of your diploid on EMM, typically ade6-M210/ade6-M216. However, because this is EMM, in the middle of your colony where the cells have run out of nutrients, the diploid will sporulate, giving a contaminatig background of azygotic asci in the colony. Therefore, your third step is to screen for diploids in your colony by picking and streaking the growing colonies from EMM onto YES. The diploids will form dark pink colonies, and will be inhibited from further sporulation by the YES. The few haploids will form lighter pink colonies which you can easily avoid picking.
Non-Sporulating Diploids
Stable, non-sporulating diploids are useful for experiments on vegetative cells that require the presence of homologous chromosomes. One example would be analysis of homologous recombination. A second might be measuring rates of chromosome loss, because diploid fission yeast that lose one of their 6 chromosomes are extremely unstable and will result in haploids. Normal diploids are so prone to sporulate, that they are impractical for these purposes because meiotic products would complicate the analysis.
Method 1
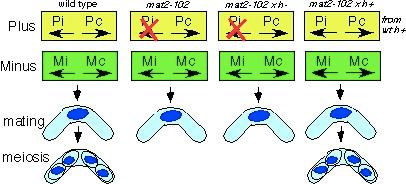
this method exploits the genetics of fission yeast mating exploiting the mat2 locus that contains the silent “plus” or P information. This information consists of two messages: Pc (for constitutive), and Pi (for induced). (See Willer et al (1995), Mol Cell Biol. 15: 4964-70). Pc is sufficient for mating but Pi is essential for proper meiosis. The mat2-102 allele eliminates the Pi information. Therefore, a homothallic (switching) strain containing mat2-102 can mate with itself, but cannot sporulate.
Like normal h90 strains, mat2-102 can mate with strains of either mating type. If it mates with an h-, it provides the (defective) plus information and cannot sporulate. However, if it mates with a wild type h+, the mat2-102 parent provides the minus information, and the partner provides intact plus information, so it can sporulate. This allows markers to be moved in and out of the mat2-102 strain: cross by h+ to makes strains, and cross by h- to isolate stable diploids. Note that because it mates with itself, you must have complementing markers to allow selection of the out-crossed zygote.
The mat2-102 locus is tightly linked to the expressed mat1 locus so can be followed easily in crosses. The logic is described in (Kohli et al, Genetics 87:471-489 (1977)) and its use in mapping is described in Bodi Z, (1991) Mol Gen Genet. 229: 77-80. Advantages the most “normal” non-sporulating diploid, with sufficient mating type heterozygosity to support a normal MI division in a pat1 meiosis. Yamamoto and Hiraoka (2003) EMBO J. 22: 2284. Disadvantages Lengthy strain constructions.
Method 2
homozygous diploids. On occasion, fission yeast will miss mitosis following S phase, and this can result in a completely homozygous diploid (endoreduplication). This strain will be homozygous for the mating type and thus unable to sporulate. These homozygous diploids can be identified by plating a strain to single colonies on YE-phloxin B and identifying the darker pink colonies (diploids). This probably occurs at 1 in 103 or less. It is imperative to verify by FACS that you have a bona-fide diploid, and not simply a spontaneous mutant that affects phloxin B staining. Advantages: easy to do for a large number of strains. Disadvantages: The strain is homozygous so the chromosomes are not differentially marked. Meiosis induced by a pat1 allele in this background lacks proper segregation in MI due to absence of mating type heterozygosity Yamamoto and Hiraoka (2003) EMBO J. 22: 2284.
Random Spore Analysis
Random spore analysis (RSA) allows many more spores to be examined than in tetrad analysis and thus facilitates rapid recombination mapping and strain construction. It’s easy to do in pombe because a simple enzyme treatment will kill remaining vegetative (unsporulated) cells and release the spores from their asci, but will have no effect on the spores themselves. However, when contemplating RSA, it is essential that all the classes of spores are viable when studying recombination frequencies, and also that double mutants may be unambiguously identified; otherwise tetrad analysis is essential. For example, it’s easy to use RSA to construct a ura- leu- double mutant, since this can be unambiguously distinguished from the ura- and leu- parental types, but it is not so straightforward to make a double temperature sensitive, unless you can reliably distinguish the phenotype of the double mutant from either parent (and if the mutants are synthetically lethal, the double mutant may be dead). There is no point in doing random spores and then having to do tetrads anyway. When in doubt, pull tetrads.
The RSA protocol is simple: Using a three day old cross check for the presence of asci under the light microscope. 1 ml of sterile distilled water is inoculated with a loopful of the cross, and glusalase (Dupont/NEN) is added to a final concentration of 0.5%. The mixture should be incubated overnight at 25-29°C or for at least 6 hours at 29°C. Glusalase is a crude snail gut enzyme that breaks down the ascus wall and kills vegetative cells. Between 200-1000 spores/plate can be plated out on YES Agar or selective medium. The plates are then incubated at an appropriate temperature until colonies form.
Random spore analysis can also be useful for plasmid complementation assays. For example, assume you have a mammalian gene on a plasmid that is homologous to a pombe gene, and you want to determine whether it cross-complements. If you have a conditional mutation in the pombe gene (e.g., temperature sensitive), you can simply transform the plasmid into the haploid strain, and raise the temperature. But more commonly, all you will have is a lethal disruption mutation. In this case, you transform your plasmid (with a different marker than the disruption) into the heterozygous diploid, sporulate, and select or screen for growth on the appropriate plates. Plasmids are usually lost at a high rate in meiosis, but this method allows you to screen a large population of spores and recover the approximately 10% that contain the plasmid. You can then determine whether those that contain the plasmid also contain the disruption marker (if it all works perfectly, about 50% of them should). Click here for considerations on building plasmids for inter-species cross-complementation experiments.
Tetrads
Tetrad analysis allows the unambiguous identification of individual meiotic products. It is an enormously powerful tool in yeast genetics, but also a bit of an art. Asci are placed in a line about 3mm apart on a YES plate using a micromanipulator (these vary from simple mechanical devices attached to a laboratory microscope, to complex microprocessor machines that automate much of the process). The ascus walls are then left to break down at 37°C for about 3-5 hours, or at 20°C overnight. Each ascus is then micromanipulated to give a line of four isolated spores, separated by about 3-5mm. The spores are incubated until colonies form at the appropriate temperature for the cross. Digestion of the ascus wall with enzymatic treatment is not necessary since the spores easily fall apart giving free spores. Also, unlike S. cerevisiae spores, S. pombe spores do not stick to each other. This means the technique of tetrad dissection is generally simpler than with S. cerevisiae. Dissecting tetrads is finicky and takes a little practice to master, but is essential to the well-trained yeast geneticist! And in this case, practice really DOES pay off.
If you are new to tetrads, you can get a free instructional information from The Singer Instrument Company in the U.K. While this is intended to help sell their micromanipulator, it also provides lots of general information that is appropriate to any micromanipulation apparatus.