Q4Q: Quantum computation for Quantum prediction of materials and molecular properties
We aim at showing that quantum computers are already useful to solve cutting-edge problems in materials science and chemistry, accessing information otherwise elusive. We pursue two objectives, making use of the two quantum computing paradigms that are currently implemented in commercial and cloud quantum hardware. Objective 1: adiabatic quantum optimization on materials databases (D-Wave). Objective 2: electronic structure of highly correlated systems on gate-model qubit arrays (IBM, Rigetti). This project is funded by the Department of Energy Office of Science, Basic Energy Sciences, under award number DE-SC0019432.

Binding mechanisms of class 2 CRISPR systems by a molecular dynamics simulations including enhanced sampling techniques
The contemporary version of genetic engineering is redefined genome editing, after the advent of molecular systems endowed with recognition properties against the DNA to be corrected. Specifically, class 2 CRISPR (clustered regularly interspaced short palindromic repeats) systems offer a unique protocol for genome editing. In order to be employed as a therapeutic tool to correct genetic imperfections or as a tool to artificially engineer crops for large-scale use, the protocol needs to be precisely sequence specific, which is still a limitation. CRISPR/Cas9 and CRISPR/Cpf1 systems share the same RNA-guided approach to cleave double-stranded DNA (dsDNA) close to the protospacer adjacent motif (PAM) that is recognized by the protein. The availability of several crystal structures of Cas9 fueled a fast development of the CRISPR/Cas9 technique. Cpf1 was harnessed for genome editing more recently, with the first demonstration in 2015. A common feature of all crystal structures is the total or partial lack of the unwound non-target DNA strand. We interrogate binding mechanisms in two class 2 CRISPR systems, with the Cas9 and Cpf1 effector proteins, by means of enhanced sampling molecular dynamics (MD) simulations, with the aim of proposing a structural/energetic framework to improve specificity. We will start from crystal structures that contain part on the unwound non-target DNA strand and use structural modeling to complete the structures. Our computational work is complemented by site-directed spin labeling (SDSL) experiments that report shape information, done in the lab of Peter Z. Qin (USC Department of Chemistry).
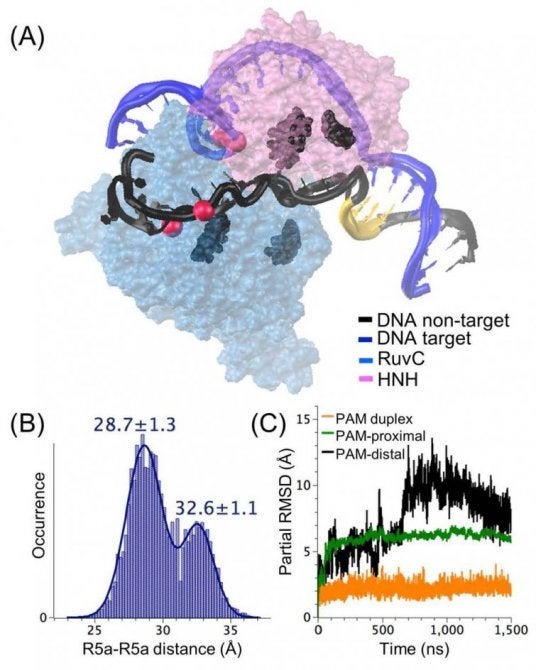
MD analysis of a Cas9 ternary complex containing the unwound DNA. (A) Overlay of the two centroid structures of the most and second-most populated clusters. For clarity, only the nuclease domains (transparent surfaces) and the DNA strands (blue = target; black = nontarget; yellow = PAM) are shown. The red spheres indicate the P atoms to which the R5a labels are attached. Representative residues in the positively charged patch (K855 and K862 in HNH, H982 and K1047 in RuvC) are highlighted. (B) Histogram of distances between R5a labels modeled at the nucleotides equivalent to c33 and n26. (C) Partial RMSD traces for segments of the DNA nontarget strand with respect to the final snapshot of the minimization−equilibration phase. Nucleotides included to represent each segment are PAM duplex, −1 to −8 downstream of PAM; PAM-proximal, +1 to +8 upstream of PAM; PAM-distal, +12 to +16 upstream of PAM. From our work: N. S. Tanprasertchai, R. Di Felice, X. Zhang, I. M. Slaymaker, C. Vazquez Reyes, W. Jian, R. Rohs, P. Z. Qin, ACS Chem. Biol. 2017, 12, 1489-1493; Angana Ray and Rosa Di Felice, “Protein-Mutation-Induced Conformational Changes of the DNA and Nuclease Domain in CRISPR/Cas9 Systems by Molecular Dynamics Simulations”, J. Phys. Chem. B 124, 2168 (2020).
We have just published our outreach article in the leading science communication publication, Scientia. https://www.scientia.global/wp-content/uploads/Rosa-Di-Felice.pdf
Fate of amyloid-β peptides on nanoparticles
The increasing technological importance of nanomaterials naturally raises the concern for possible toxic effects when they accidentally contact living organisms. Such effects will likely involve the interactions of nanomaterials with the protein arsenal of the body. On the other hand nanoparticles (NPs) have been proposed for innovative diagnostic and therapeutic approaches, applications which define the emerging field of theranostics (therapy + diagnostic) nanomedicine. In particular recent experimental works tackled the effect, in vitro, of NPs on protein fibrillation. Protein fibrillation is involved in many human diseases, including Alzheimer’s, Parkinson’s, Creutzfeld-Jacob’s and dialysis-related amyloidosis. The NPs have been found either to enhance or to inhibit the rate of formation of fibrils, therefore they can potentially lead to novel mechanisms for amyloid diseases as well as to therapeutic opportunities for their treatment. Although these results are of broad relevance, the underlying molecular mechanism (how is the NP interfering with the fibrillation process?) is poorly understood. Large scale computations have the potential to clarify such mechanism, but atomistic simulation of the problem are still in their infancy. This problem is also paradigmatic of the general field of protein-inorganic material interactions, of paramount importance in natural systems and currently a hot topic for experiments and theory.
By means of enhanced-sampling molecular dynamics (MD) simulations with atomic precision, which ensure an accurate probe of the phase space of a peptide in different environments, we recently revealed that the interaction with a solid surface stimulates the formation of elongated structures of the amyloid-β-42 (Aβ42) peptide. Along with accumulated evidence that the propensity of the single monomer to adopt fiber-like conformations is directly related to the lag time for fiber formation, we have collected key evidence that contact to a flat substrate, which may be the external surface of the cell membrane or lipoproteins, as well as foreign agents, is pivotal to fibrillation-prone structural deformations. We are now interested in assessing the role of nanoparticles of different sizes and functionalization. The rationale is that if we find suitable nanoparticles that prevent fibrillation-prone conformations, those may be used to design therapeutic strategies against Alzheimer’s disease.
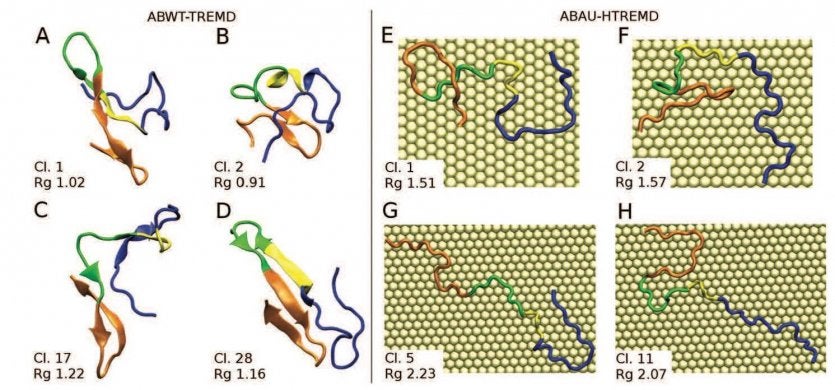
Representative conformations obtained from the cluster analysis of ABWT-TREMD (Aβ42 in solution, left) and ABAU-HTREMD (Aβ42 adsorbed on hydrated Au(111), right). Representative structures of the first (A) and second (B) most populated ABWT-TREMD clusters: in both cases the conformers show a globular shape and a Rg (radius of gyration) value lower than 1.15 nm. (C, D) Representative structures of ABWT-TREMD clusters with Rg larger than 1.15 nm (the 17th and 28th in the decreasing order of population, respectively): these are representative conformers for extended states. Representative structures of the first (E) and second (F) most populated ABAU-HTREMD clusters: these are extended states, at odds with the behavior in solution. (G, H) Representative structures of ABAU-HTREMD clusters with Rg larger than 2 nm: these are representative conformers for totally extended states. From our work: L. Bellucci, G. Bussi, R. Di Felice, S. Corni, Nanoscale2017, 9, 2279-2290.