Context
Over the last few years, parallel developments in imaging technologies, optical probes, new nanomaterials and genetic engineering have allowed the fast emergence of single molecule fluorescence techniques and their application to biological imaging. These techniques now provide unprecedented details on the spatial and temporal heterogeneities of nanoscale biological processes and are just starting to reshape our understanding of molecular organizations and interactions in cells and tissues. An emerging cell biology concept is that spatially defined subcellular compartments (e.g. immune and neuronal synapses, lipid microdomains, membrane cavities…) influence the diffusion, the location, the interactions, and thus the activity of subpopulations in an otherwise homogenous pool of the same protein. Stable nanoscale cellular compartments can thus accommodate discrete molecular interactions that deviate from the classical laws of mass action at the ensemble level, thereby allowing a few copies of a biomolecule to influence whole cell processes.
Objectives
In the Single Molecule Biophotonics Group, we aim at understanding how such discrete nanoscale interactions in plasma and nuclear membranes are capable of finely modulating normal and pathological cellular signaling and responses. For this, we design and implement ultra-sensitive imaging tools and probes that quantitatively report and influence such interactions in living cells and also in whole animals. Using highly interdisciplinary approaches, at the interface between cell and molecular biology, biological chemistry, biophysics, nanomaterial sciences and optical imaging, we seek to grasp the fundamental molecular mechanisms that govern nanostructures/protein function relationships during cellular signal processing, integration and regulation. We also aim at providing novel probes and ultra-sensitive imaging techniques for molecular diagnostics and therapeutics.
Current Projects
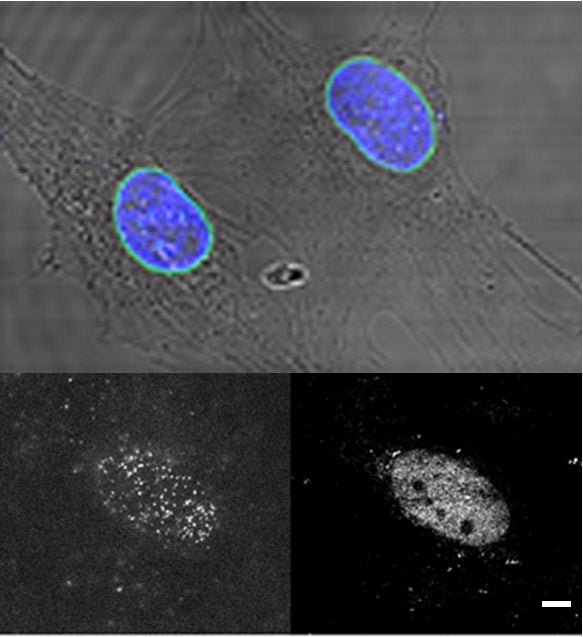
1- Nuclear membrane organization and dynamics of emerin and LINC complex components
In this project we study the nanoscale organization and the diffusion dynamics of integral proteins in the nuclear membrane of cells. At the nuclear envelope, emerin and LINC complex-associated SUN proteins interacts with the nuclear lamina meshwork to form assembly points for the mechanotransduction of cellular forces. These complexes additionally participate to transcriptional regulations and to chromatine organization, notably by tethering heterochromatin at the nuclear envelope. They contribute to the overall structural integrity of the cell nucleus by linking together the nucleoskeleton, the inner and outer nuclear membrane and the cytoskeleton and by acting as mechanotransducing centers at the nuclear envelope. Emerin and LINC-associated SUN proteins play pivotal roles in organizing these force-sensing nanohubs. Indeed, emerin or SUN protein mutations can induce muscular dystrophy diseases, which are characterized by cardiac abnormalities and skeletal muscle wasting. The molecular mechanisms by which emerin and SUN proteins protect the nuclear envelope against mechanical stress, and the reasons why their mutation induce dystrophies remain largely unknown. Using super-resolution microscopy imaging, single molecule tracking techniques and genetic tools, we study the nanoscale organization and the function of emerin and SUN proteins at the nuclear envelope with the aim of defining their contribution to cellular scaffolding, nuclear mechanics and muscular diseases.
Key words: Laminopathies, nuclear envelope, quantitative super-resolution microscopy, single molecule tracking, mechanobiology, nucleoskeletal niches, Emery-Dreifuss muscular dystophy, molecular biophysics, genetics.
Funding: National Science Foundation; Award#: 2202087; MCB program and National Institute of Health, Award#: R21AR076514; National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS).
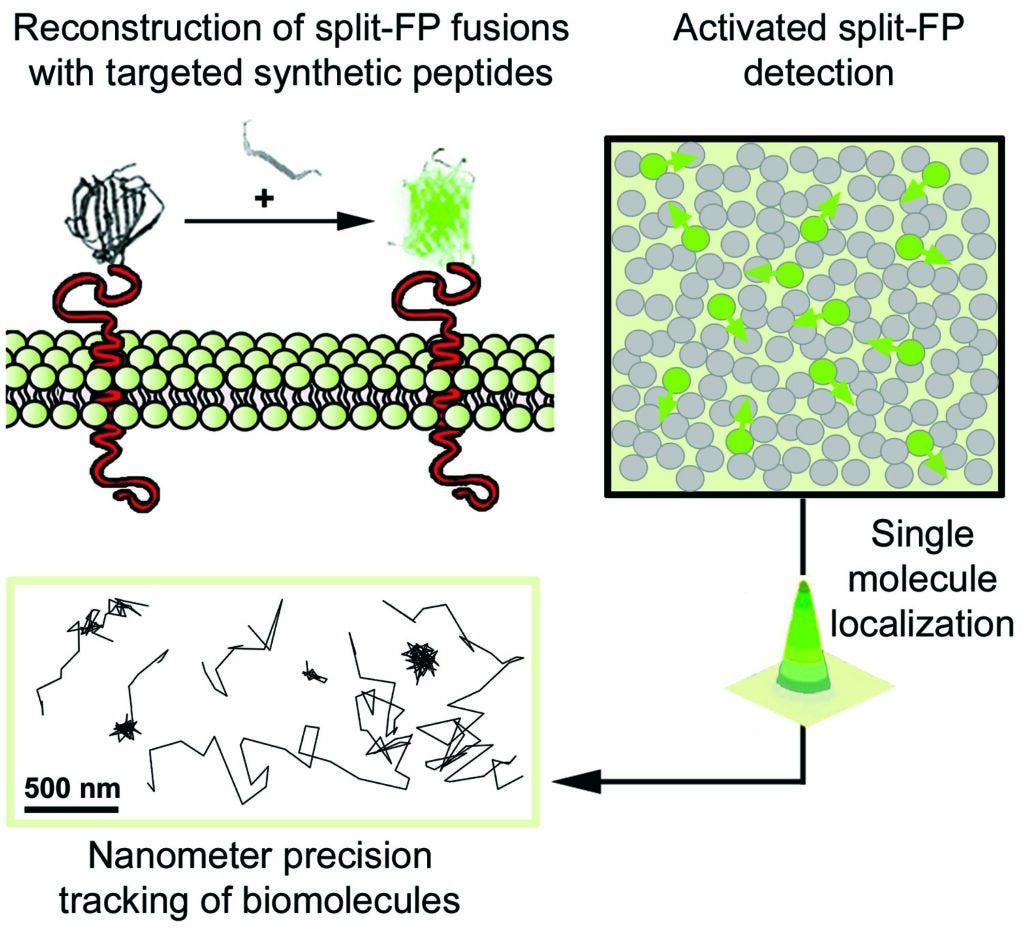
2- Nanometer-scale in vivo phenotyping of diseases by single molecule microscopy imaging in animal models
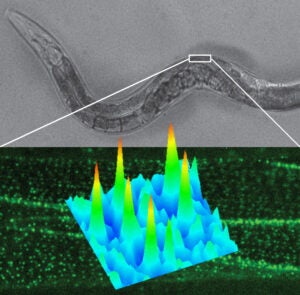
In this project we study the nanoscale distribution and the dynamics of individual membrane proteins involved in homeostatic and pathological cellular processes in live animals. For this, we use a technique called Complementation Activated Light Microscopy (CALM) and split-fluorescent protein fusions to achieve nanometer precision imaging and tracking of individual membrane receptors and channels within specific tissues inside intact living animals.
Currently, we are studying the molecular distribution and the diffusion dynamics of individual voltage-dependent Ca2+ channels within neuromuscular synapses and at the surface of muscle cells in normal and dystrophin-mutant C. elegans worm models of Duchenne muscular dystrophy. By imaging these calcium channels with nanometer precision in living worms we aim at revealing how dystrophin, a key protein involved in muscular dystrophy diseases in humans, modulates the channels’ nanoscale dynamics and activity during muscle and neuronal function.
Key words: In vivo animal imaging, Caenorhabditis elegans, single molecule detection, split-fluorescent proteins, HILO microscopy, sub-resolution CALM imaging, molecular biophysics, genetics, muscular dystrophies, calcium channels.
Funding: National Institute of Health, Award#: R21NS114911; National Institute of Neurological Disorders and Stroke (NINDS).
Past Projects
3- Structural and dynamic study of caveolin-1 membrane microdomains by single molecule imaging
At the cell plasma membrane, the scaffold protein caveolin-1 forms a variety of membrane microdomains (e.g. caveolae) that can facilitate transmembrane cross-talks between different signaling cascades by compartmentalizing signaling receptors and intracellular signal transducer proteins. Using single molecule microscopy imaging techniques, we study the cellular functions of caveolin-1 microdomains at the plasma membrane and their emerging role for signal transduction and regulation during mechanotransduction.
In particular, we employ multicolor 3D-super-resolution optical microscopy, single protein tracking and novel single molecule targeting strategies and perturbation probes to study the scaffolding and signal regulatory functions of caveolin-1 and determine their importance for the spatio-temporal modulation of mechanical signals at the nanoscale in cells.
Key words:Membrane microdomains, caveolin-1, 3D super-resolution fluorescence imaging, single molecule biophysics, cell imaging.
Funding: National Science Foundation; Award#: 1806381; Cellular Dynamics and Function program and Physics of Living Systems program.
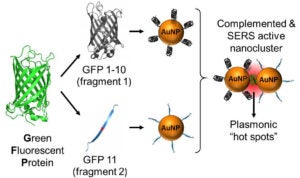
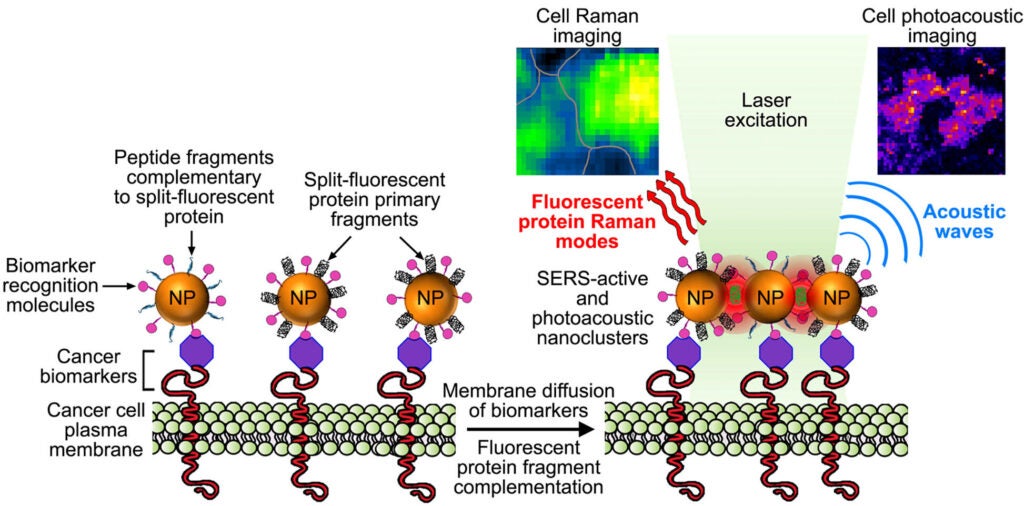
4- Activatable fluorescent protein/metal hybrid Raman nano-probes for in vitro biosensing and single cell imaging
This project involves the design and the spectroscopic characterization of a new generation of smart fluorescent protein/metal hybrid Raman nanoprobes for surface enhanced Raman scattering (SERS) biosensing and imaging. We use engineered fluorescent proteins as both molecular glue and Raman reporters to drive the self-assembly and the photonic activation of gold and silver metal nanoparticles into hot-spot SERS nanoclusters. We study the novel photonic properties and the Raman enhancement processes emerging at the protein chromophore/metal nanoparticle interfaces. These hybrid nanoprobes are further engineered to allow single molecule sensitivity detection and ultra-specific SERS biosensing both in vitro and in cells. In particular, we study the molecular factors affecting the nanoprobe directed-assembly and their activated SERS responses within live cells.
These complementary metal nanoparticles are further functionalized with targeting moities capable of recognizing and binding to overexpressed biomarkers on cancer cells. When co-targeted to tumor cells, the metal nanoparticles self-assemble into plasmonic nanoantennas. Once activated the linear nanoantennas serve as probes for highly selective detection of cancer cells over normal cells by SERS imaging of the fluorescent protein chromophore Raman signature and by photoacoustic microscopy imaging.
Key words: Nanoplasmonics, protein engineering, nanomaterial surface chemistry and assembly, SERS imaging, photoacoustic microscopy, metal nanoparticles, split-fluorescent proteins, single molecule imaging, photonics, cancer cell detection.
Funding: National Science Foundation; Award#: 1406812; Biomaterials program.