High Definition Single Cell Assay (HDSCA)
The importance of the liquid biopsy in understanding of the metastatic process and the potential use as a noninvasive route for cancer detection, characterization, and monitoring warrants the development of robust, reproducible detection and characterization of analytes for use in the clinical settings. HDSCA is our toolset to explore different phases of a disease at the single cell level. HDSCA uses the ‘No-Cell-Left-Behind’ approach to identify rare cells and characterizes them by single cell genomics and single cell proteomics. Towards that end, we have established critical academic and public-private partnerships. The ultimate goal is for our research to enable and motivate the necessary studies for clinical utility.
HDSCA has utility both as a clinical care tool and a research tool. The HDSCA workflow maintains a complete chain of custody from the patient data to the bulk sample to the individual analyte. This slide-based assay provides the most comprehensive morphometric and multi-omic analysis of circulating single cells, while the plasma from liquid biopsy samples is available for further analysis. As circulating tumor cell (CTC) assays are becoming more utilized, there is a need for development of standard operating procedures using the best pre-analytical conditions for blood collection, handling, and processing. As a member of the Blood Profiling Atlas Commons (BLOODPAC) we are making available standard operating procedures and data to the scientific community to advance the development of clinically useful tests by standardizing the methods for analysis of CTC morphology, genomics, and proteomics, with concurrent cell-free genomics (cfDNA).
The third generation HDSCA workflow provides the opportunity to identify epithelial, mesenchymal, endothelial, hematopoietic cells, as well as oncosomes (large extracellular vesicles), building a platform capable of providing a more comprehensive overview of the circulating rare events and capturing the heterogeneity of the liquid biopsy.
Click here for a video describing the approach used by HDSCA.
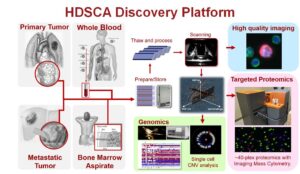
Figure: Dive et al
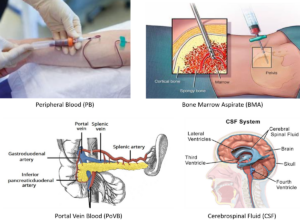
Different Types of Liquid
Different types of bodily fluids serve as important references for HDSCA due to their varying content and ability to provide a comprehensive picture of the cancer status. Here’s the different types of liquids that we explore:
- Peripheral Blood (PB) is the most commonly used liquid for analysis. It contains a wide variety of cancer-related biomarkers, including rare cells (i.e. CTCs) and acellular material (i.e. extracellular vesicles or circulating tumor DNA). Since blood circulates throughout the body, it can provide systemic information on both primary and metastatic tumors. Blood samples are relatively easy to obtain through venipuncture, making them convenient for repeated testing over time.
- Bone Marrow Aspirate (BMA) is an important reference for liquid biopsy analysis, particularly for cancers that have a high propensity to involve the bone marrow, such as prostate cancer and multiple myeloma. Bone marrow biopsies provide direct access to cancer cells in their primary or metastatic site, allowing for detailed molecular and genetic analysis, as well as a comprehensive profiling of the tumor microenvironment, including interactions between cancer cells and the bone marrow stroma.
- Portal Vein Blood (PoVB) in pancreatic cancer offers several unique advantages due to its proximity to the pancreas and the tumor microenvironment. The proximity allows for the detection of tumor-derived biomarkers at an earlier stage, potentially leading to earlier diagnosis and intervention.
- Cerebrospinal Fluid (CSF) is crucial for detecting and monitoring cancers of the brain and CNS, as it can contain CTCs, cfDNA, and other biomarkers released by these tumors. The CSF provides direct access to the tumor environment in the CNS, which is otherwise difficult to sample.