Cells are extremely sophisticated machines in which thousands of protein components and gears work together to keep the machine running. The major goal of my research is to identify small molecules that bind to certain protein gears and affect the cellular machine, thus generating new knowledge in biology and providing lead compounds for therapeutic intervention.
A central theme in my research program at USC is to exploit the strong covalent force to rapidly discover potent and specific ligands for a large number of proteins from diverse families. My lab is among the few in the world that successfully applied this strategy to the discovery of inhibitors for protein kinases, the second largest family of human proteins. Now, in collaboration with Dr. Raymond Stevens’s lab at the Bridge Institute at USC, we are the first to apply this powerful strategy to the discovery of covalent ligands for G-protein coupled receptors (GPCR), the largest family of human proteins and a major class of drug targets.
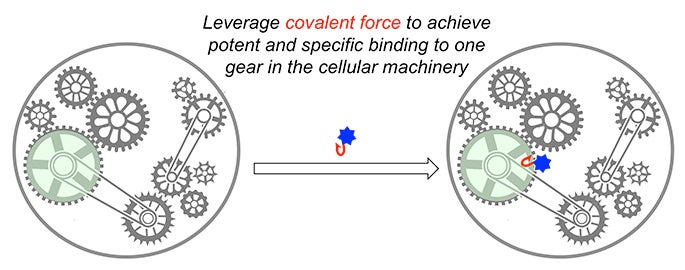
In addition, my lab recently pioneered a highly innovative methodology for discovering covalent ligands for proteins from diverse families. We were able to identify potent and specific ligands for five distinct protein targets in cells within just the past year while it would take traditional methods 3-5 years to do so for just one target. The combination of these powerful and innovative methodologies has enabled my lab to reach far beyond the boundary of conventional medicinal chemistry and create novel chemical matter to both probe biology and treat human diseases.
A number of previous and ongoing projects are described below.
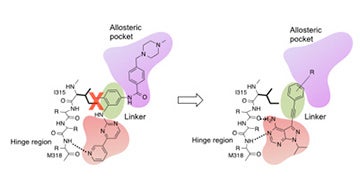
I. Structure-based drug design
Structure-based design of alkyne-containing pyrazolopyrimidines as novel inhibitors of drug-resistant Bcr-Abl kinase.
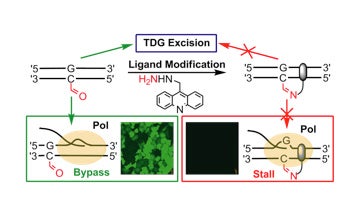
II. Development of covalent probes to monitor epigenetic modifications to DNA
This work was conducted in collaboration with Dr. Dong Wang’s lab at UC San Diego.
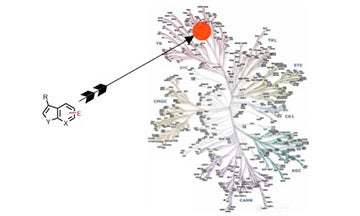
III. Development of specific, covalent inhibitors of protein kinases
Extremely potent and specific inhibitors were developed for one receptor tyrosine kinase using this strategy.
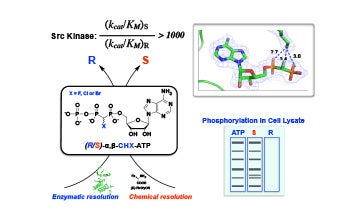
IV. Characterization of substrate analogues for enzymes
Discovered the diastereoselective utilization of novel ATP analogues by protein kinases in collaboration with Drs. Charles McKenna and Lin Chen.
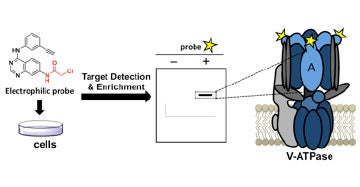
V. Identification of covalent modulators of diverse proteins
We discovered potent and highly specific covalent modulators of the vacuolar ATPase, a cellular machine that is responsible for acidifying intracellular vesicles.
Contact
Chao Zhang, Ph.D.
Associate Professor
Department of Chemistry & Loker Hydrocarbon Research Institute
University of Southern California
3430 S. Vermont Ave, TRF 114A
Los Angeles, CA 90089