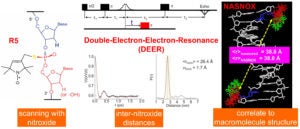
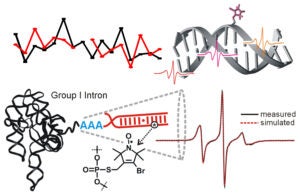
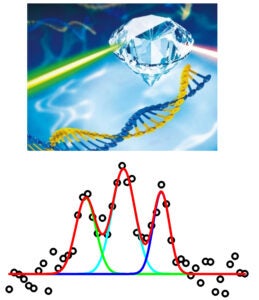
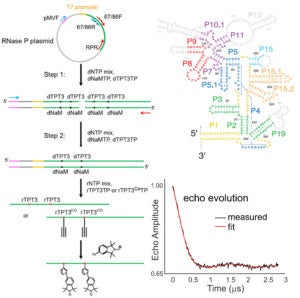
In Site-Directed Spin Labeling (SDSL), a radical with chemically stable un-paired electron(s) (e.g., a nitroxide) is attached to a specific site within a macromolecule. This yields a simple electron spin system that allows a range of problem-targeted studies. Information on the local environment at the labeling site is derived by monitoring the spin label using Electron Paramagnetic Resonance (EPR) spectroscopy. For example, continuous-wave (cw-) EPR measurements on a singly-attached nitroxide label can provide information on the steric features, polarity, and electrostatics at the labeling site. Distances between a pair of nitroxides (5 – 80 Å) can be determined using either cw- or pulsed EPR, and provide direct structural constraints.
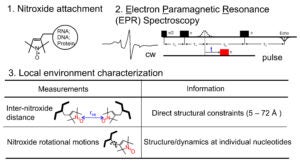 The General Strategy of Site-Directed Spin Labeling
The General Strategy of Site-Directed Spin Labeling
SDSL was initially developed for investigating structure and dynamics of membrane and membrane-associating proteins. Its scope has been expanded and has been demonstrated in large number of protein, nucleic acids, and protein-nucleic acid complexes. Several unique features of SDSL distinguish it from other methods. SDSL is capable of providing information on high molecular weight assemblies under physiological conditions using a small amount of sample (~ 5 picomoles). It is not hampered by several fundamental issues encountered in NMR (e.g., difficult to apply to large complexes) and X-ray crystallography (e.g., crystal growth; packing artifacts). Compared to Small-Angle-Xray-Scattering (SAXS) and Förster Resonance Energy Transfer (FRET), SDSL provides more precise distance measurements that better characterize conformations of the target molecule. In particular, unlike FRET that requires two chemically-distinct fluorophores, SDSL can use two chemically-identical spin labels to measure distances, which simplifies the labeling procedure. Nitroxides are smaller than most fluorophores, causing less structural perturbation. For further information, please consult references listed below.
- Hubbell, W. L., Altenbach, C. (1994) “Investigation of structure and dynamics in membrane proteins using site-directed spin labeling.” Curr Opin Struct Biol., 4, 566-73
- Sowa, Glenna Z., Qin, P. Z., (2008) “Site-directed spin labeling studies on nucleic acid structure and dynamics.” Prog. Nucleic Acids Res. Mol. Biol., 82, 147-197
- Qin, P.Z., Warncke, K. (eds.), (2015), Methods Enzymol; Vol. 563, “Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions, Part A”
- Qin, P.Z., Warncke, K. (eds.), (2015), Methods Enzymol; Vol. 564, “Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions, Part B”
The R5-Family of Nitroxide Labels
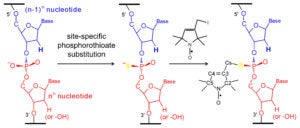
The R5-family of nitroxides are spin labels attached to phosphorothioate groups substituted at specific backbone positions of DNA and RNA. The chemistry involves two steps: (1) site-specific substitution of phosphorophioates, general achieved during solid-phase chemical synthesis of DNA or RNA; (2) post-synthetic coupling of a thio-reactive nitroxide derivatives. Because the labeling scheme takes advantage of a functional group present in all nucleotides, the R5-family of labels can be attached to arbitrary nucleotide positions to “scan” a given sequence. Studies have shown that these label can be attached with high efficiency (90% – 100%) to large DNA/RNA, and they do not severely perturb the conformation of DNA and RNA duplexes. As the cost of introducing a phosphorothioate modification is very low, and the post-synthesis nitroxide coupling can be carried out in aqueous solutions, labeling is cost-effective both in terms of money and time.
 |
 |
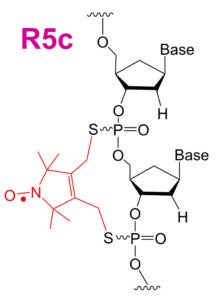 |
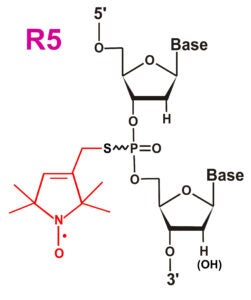
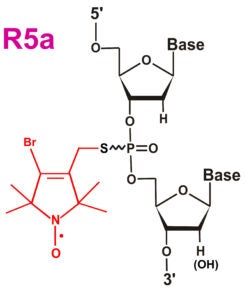
| The orignal R5 label | The R5a label with enhanced sensitivity to local steric and dynamics | The cyclic R5c label |
R5-Based Studies of Nucleic Acids and Protein-Nucleic Acid Complexes
Developments |
Applications |
|
|
Continuing Advance