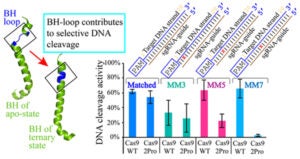
CRISPR-Cas [Clustered-Regularly-Interspaced-Short-Palindromic-Repeats (CRISPR) and CRISPR-associated (Cas) proteins] systems has ushered in a revolution in genome engineering and manipulation that is still rapidly evolving. Among the large CRISPR family, CRISPR-Cas9 and Cas12a are RNA-guided nucleases that natively cleave double-stranded DNAs at specific sites. The power of Cas9 and Cas12a lies in the programmability afforded by tunning the RNA guide sequence to form Watson-Crick pairs with a desired DNA site. However, this also subjects these enzymes to undesired off-target effects, in which erroneous binding and/or cleavage occurs on off-target genomic sequences containing imperfect RNA/DNA complementarity. Extensive mechanistic investigations have revealed that Cas9 and Cas12a interrogate a DNA target via a series of coordinated conformational changes. Further in-depth understanding on the structure-dynamics-function relationship that governs their target selection is the key to combat the off-target effect, and remains an area under active investigation.
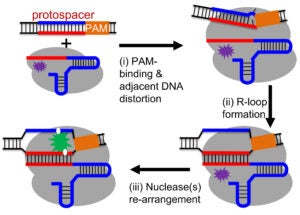
Cas9 and Cas12a Interrogate DNA Targets via Coordinated Conformational Changes
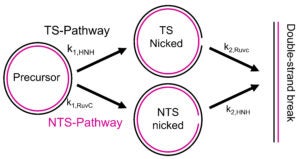
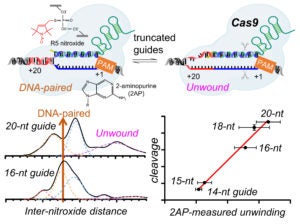
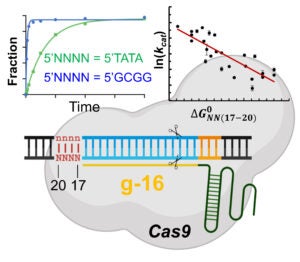
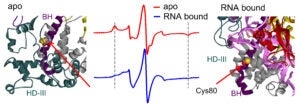
The Qin lab studies Cas9 and Cas12a mechanisms by probing structure and dynamics of on-target and off-target complexes using a combination of spin-labeling, fluorescence spectroscopy, and other biophysical methods, and correlating the finding to functional outcomes obtained with detailed enzyme kinetics analysis, bioinformatics, and other biochemical approaches. Work has uncovered key specificity check-points built upon equilibrium in DNA unwinding, and shed light on the similarity and difference between Cas9 and Cas12a. Studies are on-going on further delineating the connection between sequence-dependence DNA duplex features (DNA shape) and Cas9a/Cas12a target selection, and incorporating the finding in developing gene editing applications with enhanced specificity.
Representative Work on Cas9
Representative Work on Cas12a
 |
 |
|
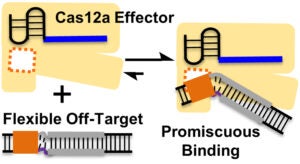
Cas12a Binds Flexible DNA Duplexes without RNA-DNA Complementarity |
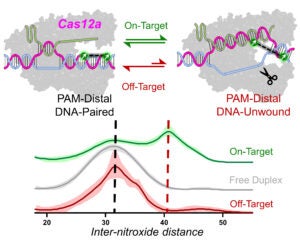
A DNA Unwinding Equilibrium Serves as a Checkpoint for Cas12a Target Discrimination |