1-D to 3-D Genomics
Since his recent selection as an Alfred P. Sloan Research Fellow, Remo Rohs continues to demonstrate the research and creativity necessary to become a leader in the scientific community.
In April and May, the assistant professor of computational biology in USC Dornsife has published five important new papers in leading scientific journals with some of these studies detailing breakthroughs in his research on protein-DNA recognition.
His findings may have profound implications for furthering research into cancer and other genetically based diseases.
Rohs’ research — which integrates two fields, genomics and structural biology — sheds light on the mechanisms underlying how proteins recognize their DNA binding sites by translating genome sequences into three-dimensional structures.
“Our research has made it possible for the first time to analyze the whole genome in terms of its three-dimensional structure,” said Rohs, who holds appointments in biological sciences, chemistry and physics in USC Dornsife and in computer science at the USC Viterbi School of Engineering. “Until now, researchers could only undertake time consuming molecular simulations for very short segments, like 20 base pairs, the building blocks of the DNA double helix.
“Now we have at hand a method that allows us to analyze the whole genome — millions of letters, very, very fast— through a computational approach we have developed.”
Rohs and his team have made this new method available to all via the DNAshape Web server based in USC Dornsife: http://rohslab.cmb.usc.edu/DNAshape/.
DNA is commonly described using its one-dimensional sequence of the letters A, C, G, and T, but as Rohs joked, proteins can’t read.
“Proteins recognize DNA through chemical and physical interactions with the three-dimensional object of the double helix,” said Rohs, referring to the structure formed by double-stranded molecules of nucleic acids. “That’s why it is important that we know the 3-D structure of DNA.”
Rohs’ team presented their methodology for the rapid prediction of DNA structural features for massive sequence data in a paper published on May 22 in Nucleic Acids Research, the second of two papers published by Rohs in the journal that month.
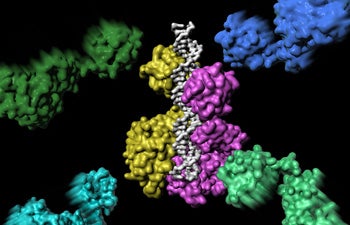
Rohs’ second Cell Reports paper was a collaboration with Xiaojiang Chen, professor of biological sciences and chemistry in USC Dornsife. The study shed light on the assembly of an important protein-DNA complex. The image illustrates how the first two of the six proteins that eventually form the complex make the initial DNA contact. Image courtesy of Ian Slaymaker.
His research will also allow scientists to visualize what happens structurally when things go wrong and disease occurs. This may have far-reaching implications for cancer research, allowing scientists to develop strategies to understand the effect of mutations in protein-DNA binding which can lead to cancer.
Rohs’ research was born out of personal dissatisfaction at not being able to look at an entire genome, but only short sequences.
“When I started my lab here at USC Dornsife three years ago, solving this problem became our first project,” Rohs said. “As we began developing a method, I understood that I needed to make this information accessible in a very fast and reliable way, so we spent a lot of effort on validating it with massive data from many different sources.”
Rohs hopes biologists will use his method to help them in their research, allowing them to visualize and analyze DNA in terms of its three-dimensional structure — a process that was previously infeasible because using molecular simulations of even a small number of base pairs could take weeks.
“More and more genome sequencing has become available as scientists all over the world have generated sequence data, but the structural field was lagging behind in making use of these data,” Rohs said. “The existing methodology was just not able to handle big amounts of sequence data. Now the methodology and the computer program we have developed will enable us to look at structural features for entire genomes for millions of sequences in minutes.”
His research is already yielding significant results.
“Remo’s powerful and unique computational tools for predicting DNA shape have enabled us to explore protein-DNA interaction from a new perspective,” said Harmen Bussemaker, associate professor in the Department of Biological Sciences at Columbia University and co-author with Rohs of a research paper on DNA methylation — a process which can be associated with cancer — published in the Proceedings of the National Academy of Sciences in April.
The tools have yielded exciting new insights into the functional impact of DNA methylation, which recently formed the basis for a successful joint grant application to the National Institutes of Health, Bussemaker added.
Rohs also published two recent papers in Cell Reports. In one, Rohs used structural information gathered from many sequences to understand for the first time how similar transcription factors from within the same family bind to different target sites in the genome in order to activate or regulate genes. He worked with Martha L. Bulyk, associate professor of medicine and pathology at Brigham & Women’s Hospital and Harvard Medical School, and a team of researchers.
“Our collaboration with Remo Rohs and his group was highly productive in investigating the influence of DNA shape on transcription factors’ DNA binding specificity,” Bulyk said. “The results of our study provide novel insights on how sequences flanking core DNA binding site motifs can lead to distinct binding preferences by closely related transcription factors.”
Rohs hopes that his analysis of rapid sequencing technology and DNA methylation data of whole genomes will reveal yet unknown molecular mechanisms underlying gene regulation.