Welcome to the Kribelbauer Lab!
Each cell in the human body contains the same genetic material, our DNA (or hardware), yet is distinct in its molecular composition. This diversity is the result of different gene expression programs defined by unique epigenetic landscapes (i.e. the software written on top of our DNA).
Transcription factors read the information encoded in our genomes by binding to specific DNA sequences, which ultimately results in epigenetic and gene expression changes. What molecular principles determine a transcription factor’s binding specificity, however, remain poorly understood.
We seek to close critical gaps in our understanding of how transcription factors modulate gene expression using high-throughput, ‘bottom-up’ genetic engineering in health and disease settings.
Areas of Focus
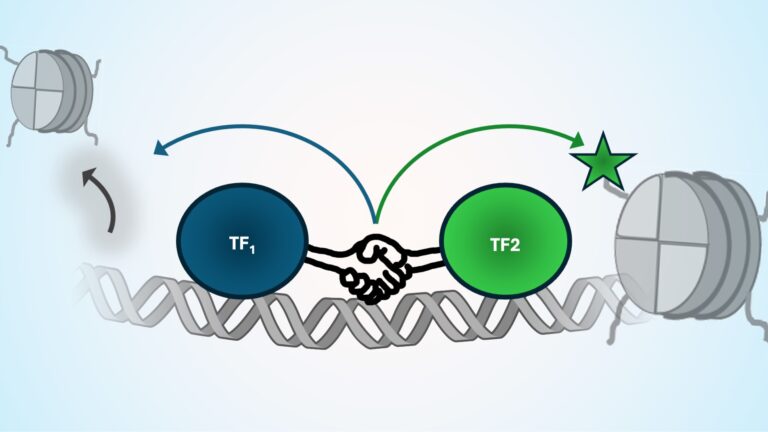
TF Cooperativity & Function
We are building new experimental tools to map TF function and cooperativity one TF-motif at a time and across a range of cellular contexts.
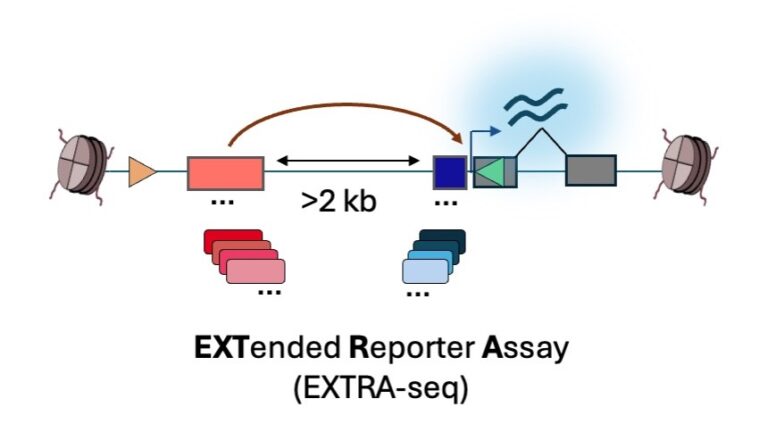
Enhancer-Promoter Communication
We are probing the compatibility rules that guide the communication of TF-bound regulatory regions (enhancers) and gene promoters using EXTRA-seq, a tool we recently developed.
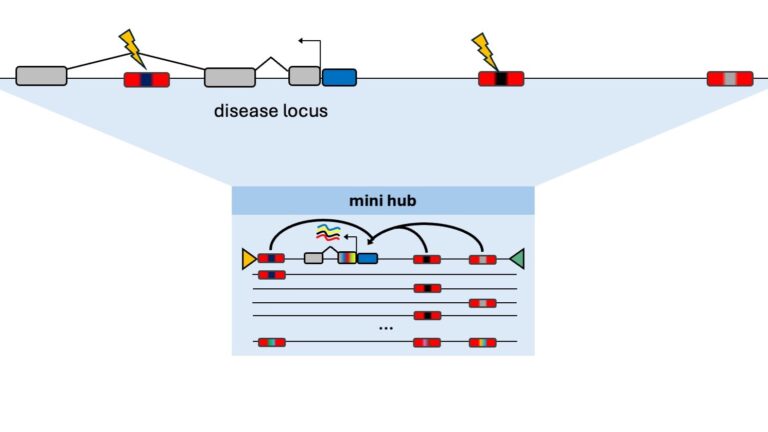
Miniaturized Gene Loci
We are building a platform to create miniaturized, disease-relevant multi-enhancer promoter modules for the high-throughput screening of deleterious expression effects caused by genetic mutations.
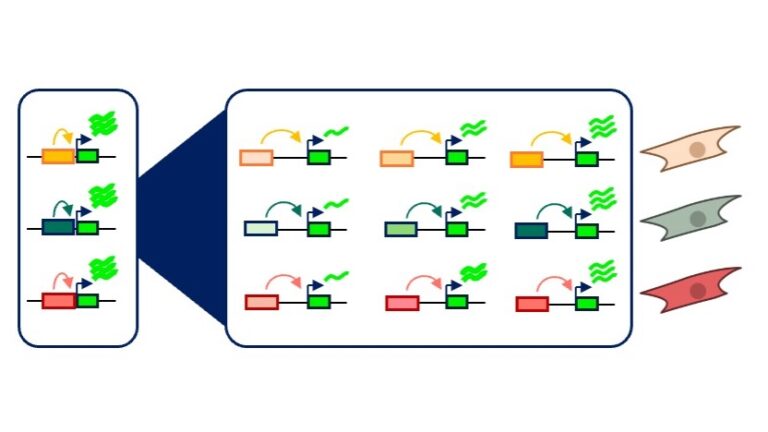
Synthetic Drivers for Gene Therapy
We strive to improve expression profiles of gene therapies by developing synthetic expression modules which are cell type-specific, tunable, and importantly, mimic native settings.
