
Comparative Functional Analyses of Mixotrophy among Microbial Eukaryotes (current project)
The ability of many single-celled, eukaryotes (protists) to combine photosynthetic and heterotrophic nutrition in a single cell is well established, but the phylogenetic breadth and tremendous ecological significance of this behavior is not widely acknowledged. ‘Mixotrophy’ is pervasive evolutionarily and ecologically. Mixotrophic behavior ranges from heterotrophic supplementation of predominantly phototrophic nutrition, to phototrophic survival of predominantly heterotrophic species under food-limited conditions, but a complex mixture of behaviors exists between these extremes. Specific benefits of heterotrophy in mixotrophic algae include the acquisition via phagotrophy or osmotrophy of carbon, energy, major nutrients (N,P) and various micronutrients (e.g. vitamins, metals, specific lipids) that provide these species with a competitive advantage over purely autotrophic species. Click here for more information.
 Studies of Marine Eukaryotic Microbes in waters of Coastal California (current project)
Studies of Marine Eukaryotic Microbes in waters of Coastal California (current project)
The study of marine Archaea and Bacteria are much further advanced than those studying more recently evolved unicellular eukaryotes. We are working with other USC collaborators, Drs. Dave Caron and John Heidelberg, to evaluate diversity and genetic complement of marine taxa from uncultured (and unstudied) eukaryotes at a time series station in the San Pedro Time Series Station off the coast of Southern California. By sequencing portions of their genomes, we can begin to evaluate gene compliment and potential function on the oceans. These data will significantly increase genomic information on environmental microbial eukaryotes in the public domain and begin to allow detailed studies of relationships between and among prokaryote and eukaryote taxa. Graduate student Amy Koid is working on this project.
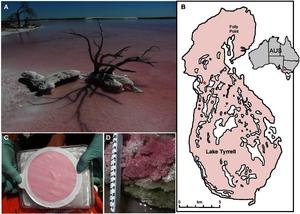 Hypersaline systems – Lake Tyrrell, AUS (past project)
Hypersaline systems – Lake Tyrrell, AUS (past project)
The Lake Tyrrell research program was in collaboration with Jill Banfield (UCB) and Eric Allen (UCSD/Scripps). Our efforts were aimed at determining the influence of extreme salinity on microbial ecosystems. To achieve this, we paired deep geomic sequencing with several other methods. Our data have provided a detailed view of how the microbial community in Lake Tyrrell is constructed and how the ecosystem functions at different salinities. The program augments the scientific foundation for understanding ecosystems response to salinization. Further infromation can be found here.

Deep Sea Hydrothermal Vent Communities (past project)
This project undertaken in collaboration with Dave Caron’s lab focuses on gene expression of marine microbial eukaryotic microbes from hydrothermal vent communities (9 North, East Pacific Rise, and Guaymas basin). See our NSF sponsored interactive educational outreach website that describes some of our activities from the 2008 cruise.

Microbial Eukaryotes in Antarctica (past project)
On a 2 month cruise in 2008 our lab took part in a collaborative cruise with Dave Caron (USC) and Darcy Longsdale (SUNY Stonybrook) in the Ross Sea to evaluate what special adaptations microbial eukaryotes have evolved to tolerate the cold, and how this affects the flow of energy in marine food chains.
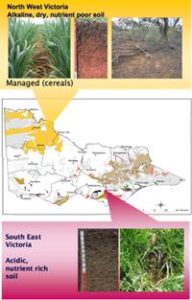 Microbial metagenomic approaches to compare agricultural soils (past project)
Microbial metagenomic approaches to compare agricultural soils (past project)
Australian soils are the most fragile in the world because they are low in organic matter, erodible and often subjected to drought. We used a metagenomic approach to inventory microbial communities from both agricultural and non-agricultural soils. Multipple sites with different soil tyepes and land use histories in SE Victoria, Australia were studied.