IMPORTANT NOTE: THE ORIGINAL COMBINATORIAL BARCODING AND INDEXING STRATEGY IS SUITABLE FOR ILLUMINA MISEQ, BUT DUE TO HIGHER INCIDENCE OF INDEX HOPPING WITH NOVASEQ AND OTHER NEW PLATFORMS WITH PATTERNED FLOWCELLS, IT IS HIGHLY RECOMMENDED TO CHANGE TO UNIQUE DUAL INDEXING (UDI). This allows informatic removal of spurious forward-reverse combinations, resembling cross-contamination between samples (up to a few percent of sequences in our experience). We recommend replacing the forward and reverse barcodes/indices with unique 8 or 10 nt index combinations. We are in the process of generating an updated protocol for using UDIs with our 515/926 primers, please contact us for more details if needed.
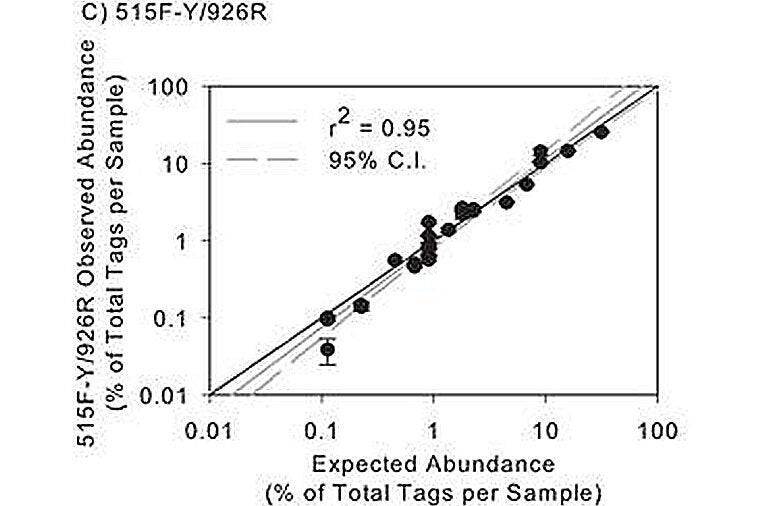
Although not widely recognized, high quality microbial community composition analysis requires periodic calibration and checking, just like any chemical assay. Small differences in PCR protocols and analytical pipelines can introduce major changes in results. Using a mock community of clones that were generated (by David Needham in our lab) to represent microbes common in mesotrophic and oligotrophic marine environments, we assessed the accuracy and precision of our 16S rRNA gene sequencing pipeline. We have selected a primer pair and analytical pipeline particularly well suited to marine and other microbiome projects. Our 515-926 primer pair not only amplifies the ssu rRNA (16S or 18S) of the vast majority of known organisms in all three domains, but testing with our prokaryotic mock communities shows that the results are remarkably quantitative. When we compared the observed vs expected abundances from our 27-member mock community, a log-log plot shows the r2 value is 0.95 (see figure to the left). This compares to an r2 ~0.5 for the popular 515-806 primers used by many labs.
Based on our results, we strongly encourage the use of a mock community for anyone using rRNA “tag sequencing,” and it is important to include the mock communities blindly within sets of samples (not run alone). Not only did we find that popular 515-806 primers poorly quantified ssu rRNA gene abundance, but it also poorly amplified members of the SAR11 cluster, the most abundant bacteria in the global surface ocean; we also found that our downstream analyses, especially clustering, were greatly informed by tracking expected vs. observed classifications and operational taxonomic unit (OTU)-generation of the mock community. Several “standard” aspects of popular analytical pipelines caused incorrect merging or splitting of mock community OTUs, but we found ways to avoid that.
Chloroplasts: Of significant note, the 16S sequences of chloroplasts in eukaryotic phytoplankton (except dinoflagellates, which have aberrant chloroplast genomes) provide a valuable assessment of their community composition, far less affected by copy number variations than more classic 18S rRNA gene sequence analysis.
For a set of recommendations on how to use the Fuhrman Lab’s mock communities and PCR primers , please see our Methods and Publications: Parada et al. 2015, Needham and Fuhrman 2016, Walters et al. 2016, Yeh et al. 2018.