Synaptic Encoding of Memories
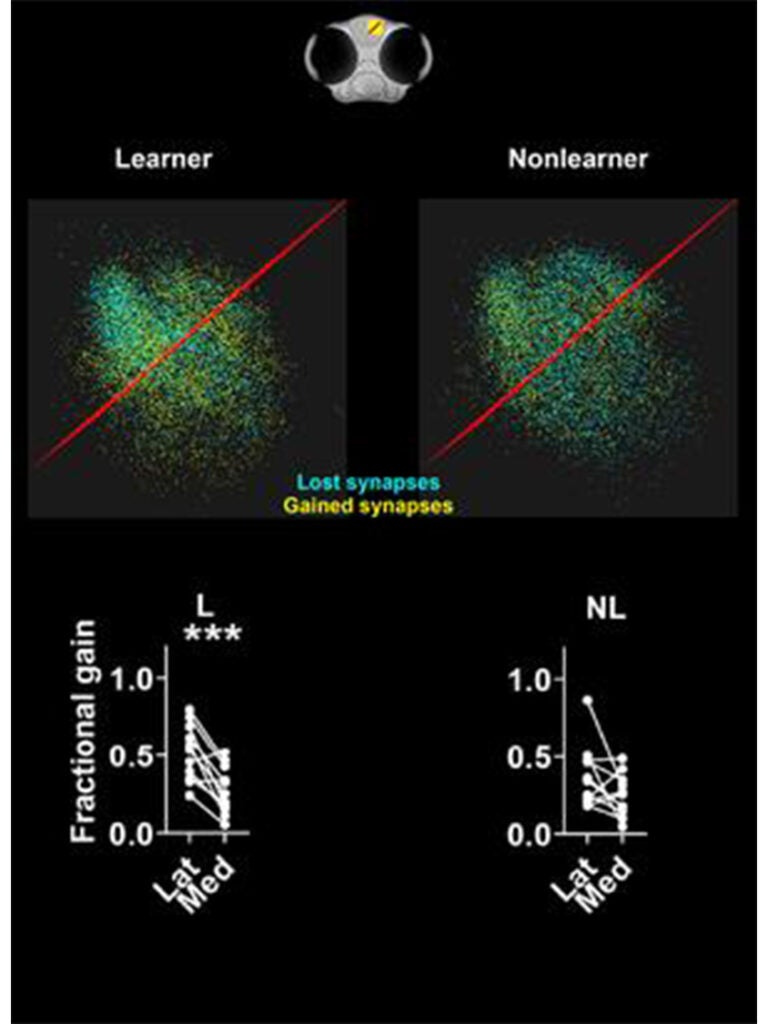
Ramon y Cajal first suggested that synapses encode memories in 1894. Since then, it has become widely accepted by neuroscientists that synapses do, in fact, represent the physical substrate of learning and memory. However, there are few, if any, examples where the spatial distribution of synapses has been mapped before and after memory formation. We developed a novel classical conditioning paradigm that caused larval zebrafish to make an associative memory. We then mapped how the distributions of synapses in the dorsal pallium changed before and after memory formation. We identified a specific region in the anterolateral dorsal pallium where synapses proliferate and an adjacent medial region where synapse loss occurs. This anterolateral region contains neurons that respond to aversive stimuli in naïve fish but, after learning, becomes responsive to the neutral stimulus that we paired with the aversive stimulus. Surprisingly, we did not see systematic changes in the strengths of existing synapses, which suggests memory formation is associated with synapse creation and elimination rather than in a change in the strength of existing synapses Dempsey (2022). We are following up this work by examining how inhibitory synapses change when an associative memory is made. We are also defining the role(s) of neurons in the anterolateral region of the pallium that respond to aversive stimuli and undergo synapse proliferation during associative memory formation.
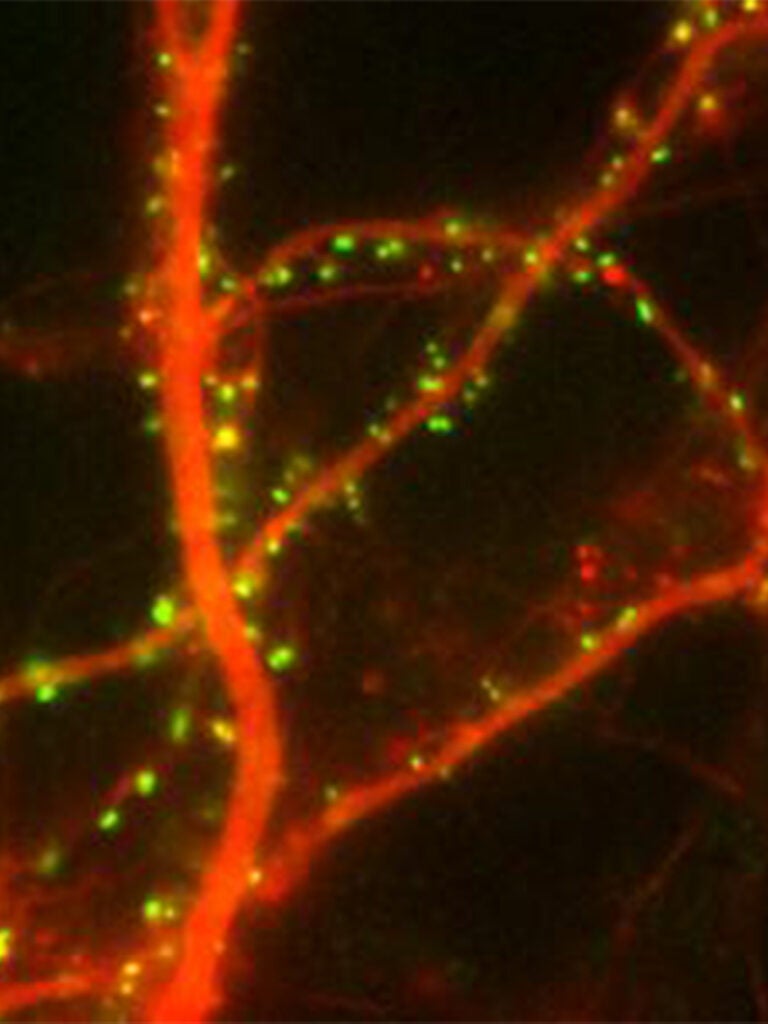
FingRs: labeling endogenous synaptic proteins in vivo
FingRs or Fibronectin intrabodies generated by mRNA display are recombinant, antibody-like proteins based on the 10FNIII Ig domain of Fibronectin that bind with very high affinity and specificity to synaptic proteins when expressed in cells in culture or in vivo. A transcriptional regulation system allows FingRs to label target synaptic proteins specifically without background. We have generated FingRs against the scaffolding proteins PSD-95 (excitatory synapses) and Gephyrin (inhibitory synapses), which can be used to map the location and strength of both types of synapses in vivo in real time Gross (2013). We have also generated a FingR against Camkii Mora (2013). Recently, we have generated mosaic transgenic zebrafish and used them to map the changes in synapses that occur when a memory is formed (see above).