The Shape of Things to Come
A USC Dornsife scientist has developed an innovative method of predicting the shape of DNA, for the first time making it expedient to do so on a genomic scale.
“DNA can have variations in shape, which are read by proteins, leading to protein-DNA binding specificity,” said Remo Rohs, assistant professor of biological sciences and chemistry, who developed the new method with his team. Protein-DNA binding specificity is the ability of a protein to recognize the nucleotide sequence of a DNA region.
Rohs used his system to aid researchers at Columbia University in executing the first systematic study of binding of a multi-protein complex to DNA. Their paper will appear in Cell this month, with Rohs as co-corresponding author.
Co-corresponding author Richard Mann, Higgins Professor of Biochemistry and Molecular Biophysics at Columbia, led a team that explored the way in which a cofactor — a protein that assists Hox proteins in performing their distinct functions — can reveal new DNA binding specificities in multiple members of the same transcription factor family. Hox proteins regulate the embryonic development of body segments across species.
For their research, the Columbia researchers needed to analyze large sections of DNA. Established methods for doing so relied on examining DNA sequence, represented by a one-dimensional string of letters, each letter corresponding to a nucleotide.
“Sequence is not enough to understand protein-DNA binding specificity,” Rohs said. “A, C, G, and T are not just letters but complex, three-dimensional objects that form the shape of the double helix.” This, in turn, makes various regions of the double helix more or less accessible to the proteins that bind to DNA.
“Bottom line, shape is important,” Rohs said. To learn more about DNA shape, Mann turned to Rohs. They had already worked together before, along with Barry Honig from Mann’s department at Columbia University, on earlier — low-throughput — shape analyses, which had also led to Cell and Nature publications.
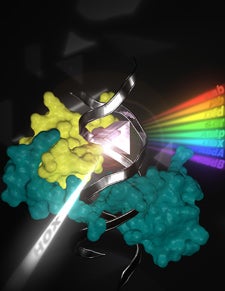
The shape of the DNA and the binding of a cofactor (yellow) unfold the binding specificities of the eight Drosophila Hox transcription factors (cyan) enabling these similar proteins to execute very distinct functions in embryonic development. Design by Ian Slaymaker.
“The initial methods for analyzing DNA shape were slow and could only analyze tens of sequences,” Mann said. “The new method Dr. Rohs developed for this paper allowed us to analyze thousands of sequences.”
The information is available “in the blink of an eye — on an ad hoc basis and for a whole genome,” Rohs said.
Rohs said his goal, also inspired by his colleagues at USC’s Molecular and Computational Biology Program, is to integrate two areas of research that have developed along parallel lines, largely disconnected from each other: sequence and structure.
While sequence research analyzes DNA in a high-throughput manner and on a genome wide basis, structure research provides three-dimensional information on DNA and proteins at atomic resolution. The new method makes this connection possible for the first time. Rohs said he hopes that DNA shape prediction will become a routine step in sequence analysis.
Rohs, who joined USC Dornsife in 2010, says that this paper was only possible because he was able to recruit a team of top-notch graduate students in his first year at USC. This research was funded by USC start-up funds, the USC-Technion Visiting Fellows Program, and an Andrew Viterbi fellowship.
Rohs is publishing a related paper that will be highlighted on the cover of ACS Chemical Biology with Tom Tullius, professor of chemistry at Boston University, examining the shape of nucleosome binding sites.